

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | NAD-dependent protein deacetylase sirtuin-1 | ||
| Ligand | BDBM50540050 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1980070 (CHEMBL4613332) | ||
| EC50 | 650±n/a nM | ||
| Citation |  Wang, Y; He, J; Liao, M; Hu, M; Li, W; Ouyang, H; Wang, X; Ye, T; Zhang, Y; Ouyang, L An overview of Sirtuins as potential therapeutic target: Structure, function and modulators. Eur J Med Chem161:48-77 (2019) [PubMed] Article Wang, Y; He, J; Liao, M; Hu, M; Li, W; Ouyang, H; Wang, X; Ye, T; Zhang, Y; Ouyang, L An overview of Sirtuins as potential therapeutic target: Structure, function and modulators. Eur J Med Chem161:48-77 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| NAD-dependent protein deacetylase sirtuin-1 | |||
| Name: | NAD-dependent protein deacetylase sirtuin-1 | ||
| Synonyms: | NAD-Dependent Deacetylase Sirtuin-1 | NAD-dependent deacetylase sirtuin 1 | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | SIR1_HUMAN | SIR2-like protein 1 | SIR2L1 | SIRT1 | Sirtuin 1 (SIRT1) | Sirtuin-1 (SIRT1) | hSIR2 | ||
| Type: | Developmental protein; hydrolase | ||
| Mol. Mass.: | 81626.66 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 747 | ||
| Sequence: |
| ||
| BDBM50540050 | |||
| n/a | |||
| Name | BDBM50540050 | ||
| Synonyms: | CHEMBL4646804 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H26F2N4O4S | ||
| Mol. Mass. | 492.539 | ||
| SMILES | Cn1nc(C(=O)N[C@H]2CC[C@H](O)CC2)c2cc(NCc3cc(F)c(F)cc3S(C)(=O)=O)ccc12 |r,wU:10.10,wD:7.6,(11.17,-18.21,;11.57,-16.73,;10.66,-15.47,;11.57,-14.22,;11.17,-12.73,;12.26,-11.64,;9.69,-12.33,;8.59,-13.42,;7.1,-13.02,;6.02,-14.11,;6.41,-15.6,;5.33,-16.69,;7.9,-16,;8.99,-14.91,;13.04,-14.7,;14.37,-13.93,;15.7,-14.69,;17.03,-13.93,;18.37,-14.69,;19.71,-13.93,;21.05,-14.7,;22.37,-13.92,;23.7,-14.7,;22.37,-12.39,;23.7,-11.62,;21.03,-11.62,;19.71,-12.39,;18.37,-11.62,;18.37,-10.08,;16.83,-11.62,;17.6,-12.95,;15.7,-16.24,;14.38,-17.01,;13.04,-16.25,)| | ||
| Structure | 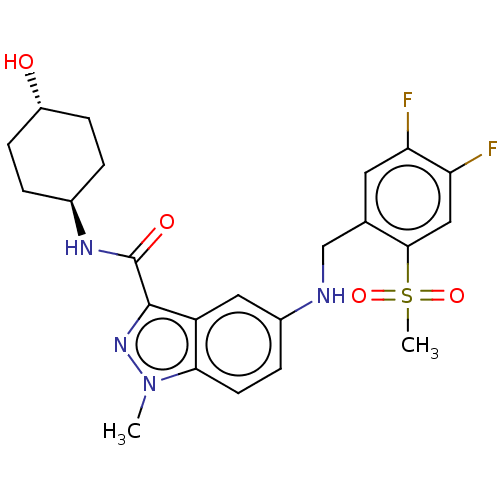
| ||