

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histone-lysine N-methyltransferase EZH2 | ||
| Ligand | BDBM50541907 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1985527 (CHEMBL4618933) | ||
| IC50 | 0.059000±n/a nM | ||
| Citation |  Khanna, A; Côté, A; Arora, S; Moine, L; Gehling, VS; Brenneman, J; Cantone, N; Stuckey, JI; Apte, S; Ramakrishnan, A; Bruderek, K; Bradley, WD; Audia, JE; Cummings, RT; Sims, RJ; Trojer, P; Levell, JR Design, Synthesis, and Pharmacological Evaluation of Second Generation EZH2 Inhibitors with Long Residence Time. ACS Med Chem Lett11:1205-1212 (2020) [PubMed] Article Khanna, A; Côté, A; Arora, S; Moine, L; Gehling, VS; Brenneman, J; Cantone, N; Stuckey, JI; Apte, S; Ramakrishnan, A; Bruderek, K; Bradley, WD; Audia, JE; Cummings, RT; Sims, RJ; Trojer, P; Levell, JR Design, Synthesis, and Pharmacological Evaluation of Second Generation EZH2 Inhibitors with Long Residence Time. ACS Med Chem Lett11:1205-1212 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histone-lysine N-methyltransferase EZH2 | |||
| Name: | Histone-lysine N-methyltransferase EZH2 | ||
| Synonyms: | ENX-1 | EZH2 | EZH2_HUMAN | Enhancer of zeste homolog 2 (EZH2) | Histone-lysine N-methyltransferase EZH2 | KMT6 | Lysine N-methyltransferase 6 | ||
| Type: | Protein | ||
| Mol. Mass.: | 85367.84 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15910 | ||
| Residue: | 746 | ||
| Sequence: |
| ||
| BDBM50541907 | |||
| n/a | |||
| Name | BDBM50541907 | ||
| Synonyms: | CHEMBL4640994 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H40N4O3S | ||
| Mol. Mass. | 536.729 | ||
| SMILES | [H][C@@]1(CC[C@@H](CC1)N1CC(C1)OC)[C@@H](C)n1c(C)c(C(=O)NCc2c(SC)cc(C)[nH]c2=O)c2ccccc12 |r,wU:4.7,1.0,wD:13.15,(41.43,-46.36,;40.68,-47.69,;39.9,-49.03,;40.67,-50.36,;42.21,-50.36,;42.98,-49.02,;42.21,-47.68,;42.98,-51.69,;42.58,-53.18,;44.07,-53.57,;44.47,-52.08,;44.84,-54.91,;44.07,-56.24,;39.9,-46.37,;40.66,-45.03,;38.36,-46.38,;37.33,-45.23,;37.66,-43.72,;35.92,-45.85,;34.59,-45.08,;34.6,-43.54,;33.26,-45.84,;31.92,-45.07,;30.59,-45.84,;30.59,-47.38,;31.92,-48.15,;31.92,-49.69,;29.25,-48.14,;27.92,-47.38,;26.59,-48.15,;27.92,-45.84,;29.25,-45.06,;29.25,-43.52,;36.09,-47.38,;35.06,-48.51,;35.52,-49.98,;37.03,-50.29,;38.06,-49.15,;37.59,-47.7,)| | ||
| Structure | 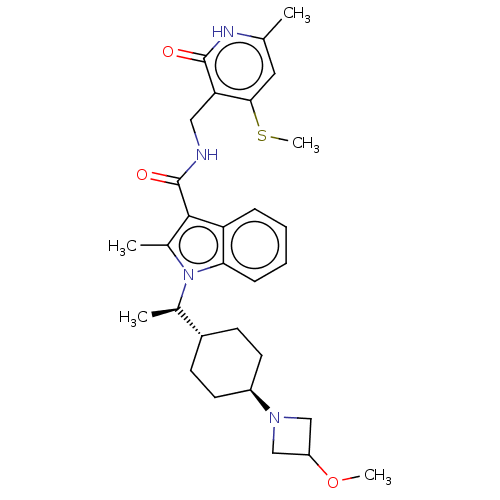
| ||