| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Ligand | BDBM50549089 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2023916 (CHEMBL4677729) |
|---|
| IC50 | 161±n/a nM |
|---|
| Citation |  Yamada, Y; Takashima, H; Walmsley, DL; Ushiyama, F; Matsuda, Y; Kanazawa, H; Yamaguchi-Sasaki, T; Tanaka-Yamamoto, N; Yamagishi, J; Kurimoto-Tsuruta, R; Ogata, Y; Ohtake, N; Angove, H; Baker, L; Harris, R; Macias, A; Robertson, A; Surgenor, A; Watanabe, H; Nakano, K; Mima, M; Iwamoto, K; Okada, A; Takata, I; Hitaka, K; Tanaka, A; Fujita, K; Sugiyama, H; Hubbard, RE Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity. J Med Chem63:14805-14820 (2020) [PubMed] Article Yamada, Y; Takashima, H; Walmsley, DL; Ushiyama, F; Matsuda, Y; Kanazawa, H; Yamaguchi-Sasaki, T; Tanaka-Yamamoto, N; Yamagishi, J; Kurimoto-Tsuruta, R; Ogata, Y; Ohtake, N; Angove, H; Baker, L; Harris, R; Macias, A; Robertson, A; Surgenor, A; Watanabe, H; Nakano, K; Mima, M; Iwamoto, K; Okada, A; Takata, I; Hitaka, K; Tanaka, A; Fujita, K; Sugiyama, H; Hubbard, RE Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity. J Med Chem63:14805-14820 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Name: | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Synonyms: | LPXC_PSEAE | Protein envA | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC) | UDP-3-O-acyl-GlcNAc deacetylase | UDP-3-O-acyl-GlcNAc deacetylase (LpxC) | envA | lpxC |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 33428.15 |
|---|
| Organism: | Pseudomonas aeruginosa |
|---|
| Description: | P47205 |
|---|
| Residue: | 303 |
|---|
| Sequence: | MIKQRTLKNIIRATGVGLHSGEKVYLTLKPAPVDTGIVFCRTDLDPVVEIPARAENVGET
TMSTTLVKGDVKVDTVEHLLSAMAGLGIDNAYVELSASEVPIMDGSAGPFVFLIQSAGLQ
EQEAAKKFIRIKREVSVEEGDKRAVFVPFDGFKVSFEIDFDHPVFRGRTQQASVDFSSTS
FVKEVSRARTFGFMRDIEYLRSQNLALGGSVENAIVVDENRVLNEDGLRYEDEFVKHKIL
DAIGDLYLLGNSLIGEFRGFKSGHALNNQLLRTLIADKDAWEVVTFEDARTAPISYMRPA
AAV
|
|
|
|---|
| BDBM50549089 |
|---|
| n/a |
|---|
| Name | BDBM50549089 |
|---|
| Synonyms: | CHEMBL4794374 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H22N2O |
|---|
| Mol. Mass. | 330.4229 |
|---|
| SMILES | CC(O)c1nccn1CCCc1ccc(cc1)C#Cc1ccccc1 |
|---|
| Structure | 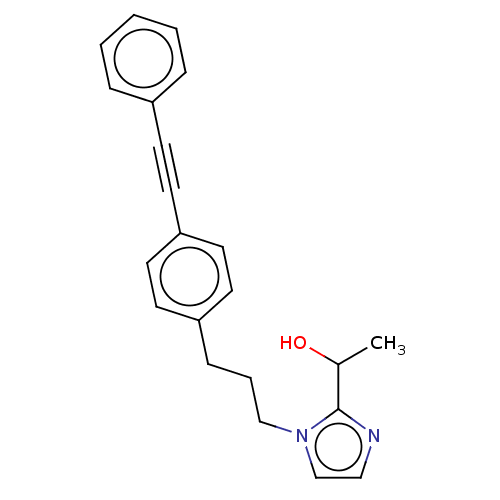
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yamada, Y; Takashima, H; Walmsley, DL; Ushiyama, F; Matsuda, Y; Kanazawa, H; Yamaguchi-Sasaki, T; Tanaka-Yamamoto, N; Yamagishi, J; Kurimoto-Tsuruta, R; Ogata, Y; Ohtake, N; Angove, H; Baker, L; Harris, R; Macias, A; Robertson, A; Surgenor, A; Watanabe, H; Nakano, K; Mima, M; Iwamoto, K; Okada, A; Takata, I; Hitaka, K; Tanaka, A; Fujita, K; Sugiyama, H; Hubbard, RE Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity. J Med Chem63:14805-14820 (2020) [PubMed] Article
Yamada, Y; Takashima, H; Walmsley, DL; Ushiyama, F; Matsuda, Y; Kanazawa, H; Yamaguchi-Sasaki, T; Tanaka-Yamamoto, N; Yamagishi, J; Kurimoto-Tsuruta, R; Ogata, Y; Ohtake, N; Angove, H; Baker, L; Harris, R; Macias, A; Robertson, A; Surgenor, A; Watanabe, H; Nakano, K; Mima, M; Iwamoto, K; Okada, A; Takata, I; Hitaka, K; Tanaka, A; Fujita, K; Sugiyama, H; Hubbard, RE Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity. J Med Chem63:14805-14820 (2020) [PubMed] Article