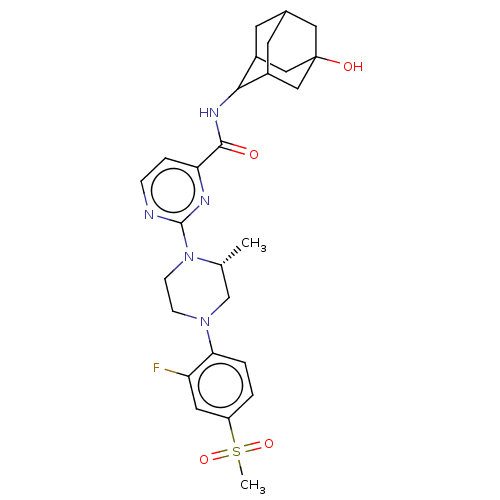
| Citation |  Ryu, JH; Lee, JA; Kim, S; Shin, YA; Yang, J; Han, HY; Son, HJ; Kim, YH; Sa, JH; Kim, JS; Lee, J; Lee, J; Park, HG Discovery of 2-((R)-4-(2-Fluoro-4-(methylsulfonyl)phenyl)-2-methylpiperazin-1-yl)-N-((1R,2s,3S,5S,7S)-5-hydroxyadamantan-2-yl)pyrimidine-4-carboxamide (SKI2852): A Highly Potent, Selective, and Orally Bioavailable Inhibitor of 11?-Hydroxysteroid Dehydrogenase Type 1 (11?-HSD1). J Med Chem59:10176-10189 (2016) [PubMed] Ryu, JH; Lee, JA; Kim, S; Shin, YA; Yang, J; Han, HY; Son, HJ; Kim, YH; Sa, JH; Kim, JS; Lee, J; Lee, J; Park, HG Discovery of 2-((R)-4-(2-Fluoro-4-(methylsulfonyl)phenyl)-2-methylpiperazin-1-yl)-N-((1R,2s,3S,5S,7S)-5-hydroxyadamantan-2-yl)pyrimidine-4-carboxamide (SKI2852): A Highly Potent, Selective, and Orally Bioavailable Inhibitor of 11?-Hydroxysteroid Dehydrogenase Type 1 (11?-HSD1). J Med Chem59:10176-10189 (2016) [PubMed] |
|---|
| SMILES | C[C@@H]1CN(CCN1c1nccc(n1)C(=O)NC1C2CC3CC1CC(O)(C3)C2)c1ccc(cc1F)S(C)(=O)=O |r,wD:1.0,TLB:15:16:25.19.20:22,16:17:25:20.21.22,THB:18:19:22:26.17.16,18:17:25.19.20:22,16:21:25:26.18.17,(37.25,-23.6,;37.25,-22.06,;35.92,-21.28,;35.92,-19.74,;37.26,-18.98,;38.59,-19.75,;38.58,-21.29,;39.91,-22.07,;39.91,-23.61,;41.25,-24.38,;42.58,-23.61,;42.58,-22.06,;41.24,-21.3,;43.91,-21.28,;43.9,-19.74,;45.25,-22.05,;46.58,-21.27,;46.55,-19.75,;47.53,-18.44,;48.95,-18.98,;48.99,-20.56,;47.99,-21.82,;49.32,-21.31,;49.28,-19.82,;50.75,-20.21,;50.44,-18.52,;47.94,-19.37,;34.6,-18.97,;34.6,-17.43,;33.27,-16.65,;31.94,-17.42,;31.94,-18.97,;33.27,-19.73,;33.26,-21.27,;30.6,-16.64,;29.27,-17.41,;29.82,-15.3,;31.37,-15.3,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ryu, JH; Lee, JA; Kim, S; Shin, YA; Yang, J; Han, HY; Son, HJ; Kim, YH; Sa, JH; Kim, JS; Lee, J; Lee, J; Park, HG Discovery of 2-((R)-4-(2-Fluoro-4-(methylsulfonyl)phenyl)-2-methylpiperazin-1-yl)-N-((1R,2s,3S,5S,7S)-5-hydroxyadamantan-2-yl)pyrimidine-4-carboxamide (SKI2852): A Highly Potent, Selective, and Orally Bioavailable Inhibitor of 11?-Hydroxysteroid Dehydrogenase Type 1 (11?-HSD1). J Med Chem59:10176-10189 (2016) [PubMed]
Ryu, JH; Lee, JA; Kim, S; Shin, YA; Yang, J; Han, HY; Son, HJ; Kim, YH; Sa, JH; Kim, JS; Lee, J; Lee, J; Park, HG Discovery of 2-((R)-4-(2-Fluoro-4-(methylsulfonyl)phenyl)-2-methylpiperazin-1-yl)-N-((1R,2s,3S,5S,7S)-5-hydroxyadamantan-2-yl)pyrimidine-4-carboxamide (SKI2852): A Highly Potent, Selective, and Orally Bioavailable Inhibitor of 11?-Hydroxysteroid Dehydrogenase Type 1 (11?-HSD1). J Med Chem59:10176-10189 (2016) [PubMed]