| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM50125242 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212863 (CHEMBL878974) |
|---|
| Ki | >4200±n/a nM |
|---|
| Citation |  Quan, ML; Ellis, CD; He, MY; Liauw, AY; Lam, PY; Rossi, KA; Knabb, RM; Luettgen, JM; Wright, MR; Wong, PC; Wexler, RR Nonbenzamidine isoxazoline derivatives as factor Xa inhibitors. Bioorg Med Chem Lett13:1023-8 (2003) [PubMed] Quan, ML; Ellis, CD; He, MY; Liauw, AY; Lam, PY; Rossi, KA; Knabb, RM; Luettgen, JM; Wright, MR; Wong, PC; Wexler, RR Nonbenzamidine isoxazoline derivatives as factor Xa inhibitors. Bioorg Med Chem Lett13:1023-8 (2003) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Alpha-trypsin chain 1 | Alpha-trypsin chain 2 | Beta-trypsin | Cationic trypsinogen | PRSS1 | Serine protease 1 | TRP1 | TRY1 | TRY1_HUMAN | TRYP1 | Thrombin & trypsin | Trypsin | Trypsin I | Trypsin-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 26557.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07477 |
|---|
| Residue: | 247 |
|---|
| Sequence: | MNPLLILTFVAAALAAPFDDDDKIVGGYNCEENSVPYQVSLNSGYHFCGGSLINEQWVVS
AGHCYKSRIQVRLGEHNIEVLEGNEQFINAAKIIRHPQYDRKTLNNDIMLIKLSSRAVIN
ARVSTISLPTAPPATGTKCLISGWGNTASSGADYPDELQCLDAPVLSQAKCEASYPGKIT
SNMFCVGFLEGGKDSCQGDSGGPVVCNGQLQGVVSWGDGCAQKNKPGVYTKVYNYVKWIK
NTIAANS
|
|
|
|---|
| BDBM50125242 |
|---|
| n/a |
|---|
| Name | BDBM50125242 |
|---|
| Synonyms: | 3-(3-Amino-4-chloro-phenyl)-5-(methanesulfonylamino-methyl)-4,5-dihydro-isoxazole-5-carboxylic acid [5-(2-sulfamoyl-phenyl)-pyridin-2-yl]-amide | CHEMBL7626 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H23ClN6O6S2 |
|---|
| Mol. Mass. | 579.048 |
|---|
| SMILES | CS(=O)(=O)NCC1(CC(=NO1)c1ccc(Cl)c(N)c1)C(=O)Nc1ccc(cn1)-c1ccccc1S(N)(=O)=O |c:8| |
|---|
| Structure | 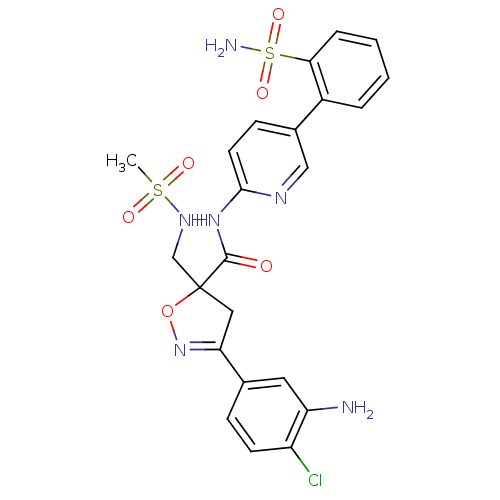
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Quan, ML; Ellis, CD; He, MY; Liauw, AY; Lam, PY; Rossi, KA; Knabb, RM; Luettgen, JM; Wright, MR; Wong, PC; Wexler, RR Nonbenzamidine isoxazoline derivatives as factor Xa inhibitors. Bioorg Med Chem Lett13:1023-8 (2003) [PubMed]
Quan, ML; Ellis, CD; He, MY; Liauw, AY; Lam, PY; Rossi, KA; Knabb, RM; Luettgen, JM; Wright, MR; Wong, PC; Wexler, RR Nonbenzamidine isoxazoline derivatives as factor Xa inhibitors. Bioorg Med Chem Lett13:1023-8 (2003) [PubMed]