| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Indoleamine 2,3-dioxygenase 1 |
|---|
| Ligand | BDBM50567132 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2102391 (CHEMBL4810787) |
|---|
| IC50 | 0.100000±n/a nM |
|---|
| Citation |  Yu, W; Deng, Y; Sloman, D; Li, D; Liu, K; Fradera, X; Lesburg, CA; Martinot, T; Doty, A; Ferguson, H; Richard Miller, J; Knemeyer, I; Otte, K; Vincent, S; Sciammetta, N; Jonathan Bennett, D; Han, Y Discovery of IDO1 inhibitors containing a decahydroquinoline, decahydro-1,6-naphthyridine, or octahydro-1H-pyrrolo[3,2-c]pyridine scaffold. Bioorg Med Chem Lett49:0 (2021) [PubMed] Article Yu, W; Deng, Y; Sloman, D; Li, D; Liu, K; Fradera, X; Lesburg, CA; Martinot, T; Doty, A; Ferguson, H; Richard Miller, J; Knemeyer, I; Otte, K; Vincent, S; Sciammetta, N; Jonathan Bennett, D; Han, Y Discovery of IDO1 inhibitors containing a decahydroquinoline, decahydro-1,6-naphthyridine, or octahydro-1H-pyrrolo[3,2-c]pyridine scaffold. Bioorg Med Chem Lett49:0 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Indoleamine 2,3-dioxygenase 1 |
|---|
| Name: | Indoleamine 2,3-dioxygenase 1 |
|---|
| Synonyms: | I23O1_HUMAN | IDO | IDO-1 | IDO1 | INDO | Indoleamine 2,3-Dioxygenasae (IDO) | Indoleamine 2,3-dioxygenase | Indoleamine-pyrrole 2,3-dioxygenase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 45330.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14902 |
|---|
| Residue: | 403 |
|---|
| Sequence: | MAHAMENSWTISKEYHIDEEVGFALPNPQENLPDFYNDWMFIAKHLPDLIESGQLRERVE
KLNMLSIDHLTDHKSQRLARLVLGCITMAYVWGKGHGDVRKVLPRNIAVPYCQLSKKLEL
PPILVYADCVLANWKKKDPNKPLTYENMDVLFSFRDGDCSKGFFLVSLLVEIAAASAIKV
IPTVFKAMQMQERDTLLKALLEIASCLEKALQVFHQIHDHVNPKAFFSVLRIYLSGWKGN
PQLSDGLVYEGFWEDPKEFAGGSAGQSSVFQCFDVLLGIQQTAGGGHAAQFLQDMRRYMP
PAHRNFLCSLESNPSVREFVLSKGDAGLREAYDACVKALVSLRSYHLQIVTKYILIPASQ
QPKENKTSEDPSKLEAKGTGGTDLMNFLKTVRSTTEKSLLKEG
|
|
|
|---|
| BDBM50567132 |
|---|
| n/a |
|---|
| Name | BDBM50567132 |
|---|
| Synonyms: | CHEMBL4863767 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H28F4N4O |
|---|
| Mol. Mass. | 476.5096 |
|---|
| SMILES | Fc1ccc(NC(=O)C2(CCC2)C2CCC3C(CCCN3c3ccnc(n3)C(F)(F)F)C2)cc1 |
|---|
| Structure | 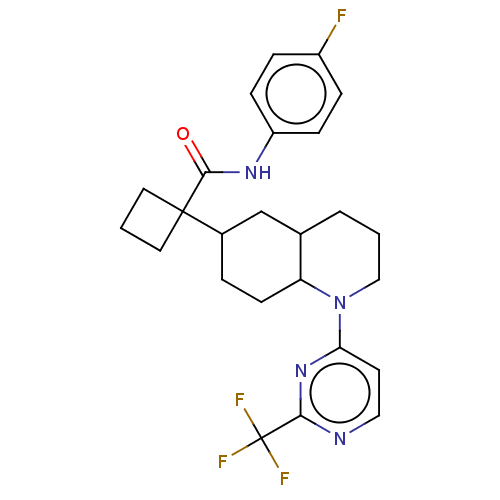
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yu, W; Deng, Y; Sloman, D; Li, D; Liu, K; Fradera, X; Lesburg, CA; Martinot, T; Doty, A; Ferguson, H; Richard Miller, J; Knemeyer, I; Otte, K; Vincent, S; Sciammetta, N; Jonathan Bennett, D; Han, Y Discovery of IDO1 inhibitors containing a decahydroquinoline, decahydro-1,6-naphthyridine, or octahydro-1H-pyrrolo[3,2-c]pyridine scaffold. Bioorg Med Chem Lett49:0 (2021) [PubMed] Article
Yu, W; Deng, Y; Sloman, D; Li, D; Liu, K; Fradera, X; Lesburg, CA; Martinot, T; Doty, A; Ferguson, H; Richard Miller, J; Knemeyer, I; Otte, K; Vincent, S; Sciammetta, N; Jonathan Bennett, D; Han, Y Discovery of IDO1 inhibitors containing a decahydroquinoline, decahydro-1,6-naphthyridine, or octahydro-1H-pyrrolo[3,2-c]pyridine scaffold. Bioorg Med Chem Lett49:0 (2021) [PubMed] Article