

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Melanocortin receptor 4 | ||
| Ligand | BDBM50189022 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_395996 (CHEMBL910341) | ||
| Ki | 5±n/a nM | ||
| Citation |  Tian, X; Mishra, RK; Switzer, AG; Hu, XE; Kim, N; Mazur, AW; Ebetino, FH; Wos, JA; Crossdoersen, D; Pinney, BB; Farmer, JA; Sheldon, RJ Design and synthesis of potent and selective 1,3,4-trisubstituted-2-oxopiperazine based melanocortin-4 receptor agonists. Bioorg Med Chem Lett16:4668-73 (2006) [PubMed] Article Tian, X; Mishra, RK; Switzer, AG; Hu, XE; Kim, N; Mazur, AW; Ebetino, FH; Wos, JA; Crossdoersen, D; Pinney, BB; Farmer, JA; Sheldon, RJ Design and synthesis of potent and selective 1,3,4-trisubstituted-2-oxopiperazine based melanocortin-4 receptor agonists. Bioorg Med Chem Lett16:4668-73 (2006) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Melanocortin receptor 4 | |||
| Name: | Melanocortin receptor 4 | ||
| Synonyms: | MC4-R | MC4R | MC4R_HUMAN | Melanocortin MC4 | Melanocortin receptor 4 (MC-4) | Melanocortin receptor 4 (MC4-R) | Melanocortin receptor 4 (MC4R) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 36949.50 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P32245 | ||
| Residue: | 332 | ||
| Sequence: |
| ||
| BDBM50189022 | |||
| n/a | |||
| Name | BDBM50189022 | ||
| Synonyms: | (S)-2-[(S)-4-[(R)-2-[(S)-2-acetylamino-3-(3H-imidazol-4-yl)-propionylamino]-3-(4-fluoro-phenyl)-propionyl]-3-(3-guanidino-propyl)-2-oxo-piperazin-1-yl]-N-methyl-3-naphthalen-2-yl-propionamide | CHEMBL384774 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C39H47FN10O5 | ||
| Mol. Mass. | 754.8529 | ||
| SMILES | CNC(=O)[C@H](Cc1ccc2ccccc2c1)N1CCN([C@@H](CCCN=C(N)N)C1=O)C(=O)[C@@H](Cc1ccc(F)cc1)NC(=O)[C@H](Cc1cnc[nH]1)NC(C)=O |wU:20.22,4.17,32.44,44.55,(15.61,-28.99,;16.95,-28.22,;18.29,-29,;18.28,-30.54,;19.62,-28.23,;20.95,-29.01,;20.95,-30.55,;19.61,-31.31,;19.61,-32.85,;20.94,-33.61,;20.94,-35.15,;22.28,-35.93,;23.61,-35.14,;23.6,-33.61,;22.28,-32.85,;22.28,-31.31,;19.63,-26.68,;18.29,-25.92,;18.3,-24.38,;19.63,-23.6,;20.97,-24.38,;22.3,-23.6,;23.63,-24.37,;24.96,-23.6,;26.29,-24.37,;27.63,-23.59,;28.97,-24.36,;27.63,-22.05,;20.96,-25.92,;22.29,-26.69,;19.63,-22.06,;20.96,-21.31,;18.29,-21.31,;16.95,-22.07,;15.62,-21.31,;14.28,-22.07,;12.95,-21.31,;12.95,-19.77,;11.61,-19.01,;14.28,-18.99,;15.61,-19.76,;18.29,-19.77,;19.63,-18.99,;20.97,-19.77,;19.63,-17.45,;20.97,-16.68,;22.31,-17.45,;22.47,-18.98,;23.98,-19.31,;24.76,-17.97,;23.72,-16.82,;18.29,-16.68,;18.29,-15.14,;16.95,-14.37,;19.62,-14.36,)| | ||
| Structure | 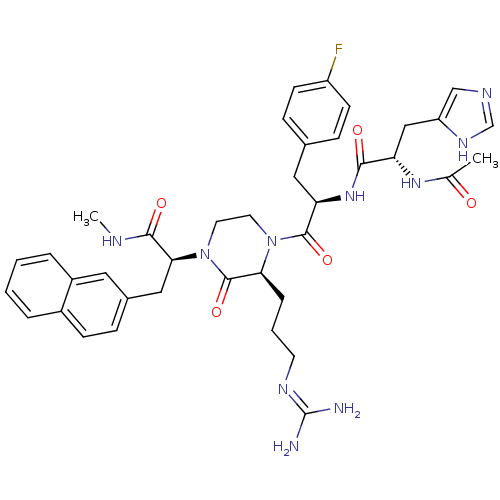
| ||