| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM50606572 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2260540 (CHEMBL5215551) |
|---|
| IC50 | <0.300000±n/a nM |
|---|
| Citation |  Yu, W; Liu, J; Clausen, D; Yu, Y; Duffy, JL; Wang, M; Xu, S; Deng, L; Suzuki, T; Chung, CC; Myers, RW; Klein, DJ; Fells, JI; Holloway, MK; Wu, J; Wu, G; Howell, BJ; Barnard, RJO; Kozlowski, J Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity. J Med Chem64:4709-4729 (2021) [PubMed] Article Yu, W; Liu, J; Clausen, D; Yu, Y; Duffy, JL; Wang, M; Xu, S; Deng, L; Suzuki, T; Chung, CC; Myers, RW; Klein, DJ; Fells, JI; Holloway, MK; Wu, J; Wu, G; Howell, BJ; Barnard, RJO; Kozlowski, J Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity. J Med Chem64:4709-4729 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM50606572 |
|---|
| n/a |
|---|
| Name | BDBM50606572 |
|---|
| Synonyms: | CHEMBL5218693 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C33H44N4O4 |
|---|
| Mol. Mass. | 560.7269 |
|---|
| SMILES | CCN1CCC2(C[C@@H]2C(=O)N[C@@H](CCCCCC(=O)CC)c2ncc(o2)-c2cc3ccc(C)nc3cc2OC)CC1 |r| |
|---|
| Structure | 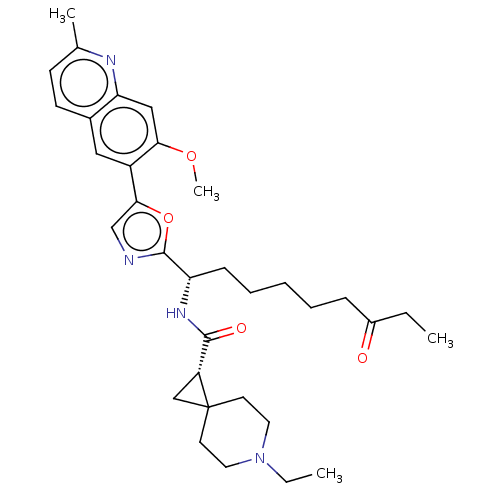
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yu, W; Liu, J; Clausen, D; Yu, Y; Duffy, JL; Wang, M; Xu, S; Deng, L; Suzuki, T; Chung, CC; Myers, RW; Klein, DJ; Fells, JI; Holloway, MK; Wu, J; Wu, G; Howell, BJ; Barnard, RJO; Kozlowski, J Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity. J Med Chem64:4709-4729 (2021) [PubMed] Article
Yu, W; Liu, J; Clausen, D; Yu, Y; Duffy, JL; Wang, M; Xu, S; Deng, L; Suzuki, T; Chung, CC; Myers, RW; Klein, DJ; Fells, JI; Holloway, MK; Wu, J; Wu, G; Howell, BJ; Barnard, RJO; Kozlowski, J Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity. J Med Chem64:4709-4729 (2021) [PubMed] Article