| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein phosphatase non-receptor type 1 |
|---|
| Ligand | BDBM50189668 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_368413 (CHEMBL870503) |
|---|
| IC50 | 240±n/a nM |
|---|
| Citation |  Combs, AP; Zhu, W; Crawley, ML; Glass, B; Polam, P; Sparks, RB; Modi, D; Takvorian, A; McLaughlin, E; Yue, EW; Wasserman, Z; Bower, M; Wei, M; Rupar, M; Ala, PJ; Reid, BM; Ellis, D; Gonneville, L; Emm, T; Taylor, N; Yeleswaram, S; Li, Y; Wynn, R; Burn, TC; Hollis, G; Liu, PC; Metcalf, B Potent benzimidazole sulfonamide protein tyrosine phosphatase 1B inhibitors containing the heterocyclic (S)-isothiazolidinone phosphotyrosine mimetic. J Med Chem49:3774-89 (2006) [PubMed] Article Combs, AP; Zhu, W; Crawley, ML; Glass, B; Polam, P; Sparks, RB; Modi, D; Takvorian, A; McLaughlin, E; Yue, EW; Wasserman, Z; Bower, M; Wei, M; Rupar, M; Ala, PJ; Reid, BM; Ellis, D; Gonneville, L; Emm, T; Taylor, N; Yeleswaram, S; Li, Y; Wynn, R; Burn, TC; Hollis, G; Liu, PC; Metcalf, B Potent benzimidazole sulfonamide protein tyrosine phosphatase 1B inhibitors containing the heterocyclic (S)-isothiazolidinone phosphotyrosine mimetic. J Med Chem49:3774-89 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein phosphatase non-receptor type 1 |
|---|
| Name: | Tyrosine-protein phosphatase non-receptor type 1 |
|---|
| Synonyms: | PTN1_HUMAN | PTP1B | PTPN1 | Protein tyrosine phosphatase 1B (PTP1B) | Protein tyrosine phosphatase-1B (PTP1B) | Protein-tyrosine phosphatase 1B | Protein-tyrosine phosphatase 1B (PTP1B) | Tyrosine-protein phosphatase non-receptor type 1 | Tyrosine-protein phosphatase non-receptor type 1 (PTP1B) |
|---|
| Type: | Protein phosphatase |
|---|
| Mol. Mass.: | 49963.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human recombinant GST-fusion PTP1B (1-435). |
|---|
| Residue: | 435 |
|---|
| Sequence: | MEMEKEFEQIDKSGSWAAIYQDIRHEASDFPCRVAKLPKNKNRNRYRDVSPFDHSRIKLH
QEDNDYINASLIKMEEAQRSYILTQGPLPNTCGHFWEMVWEQKSRGVVMLNRVMEKGSLK
CAQYWPQKEEKEMIFEDTNLKLTLISEDIKSYYTVRQLELENLTTQETREILHFHYTTWP
DFGVPESPASFLNFLFKVRESGSLSPEHGPVVVHCSAGIGRSGTFCLADTCLLLMDKRKD
PSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLE
PPPEHIPPPPRPPKRILEPHNGKCREFFPNHQWVKEETQEDKDCPIKEEKGSPLNAAPYG
IESMSQDTEVRSRVVGGSLRGAQAASPAKGEPSLPEKDEDHALSYWKPFLVNMCVATVLT
AGAYLCYRFLFNSNT
|
|
|
|---|
| BDBM50189668 |
|---|
| n/a |
|---|
| Name | BDBM50189668 |
|---|
| Synonyms: | CHEMBL208984 | N-{1-(1H-benzoimidazol-2-yl)-2-[4-(1,1,3-trioxo-1lambda*6*-isothiazolidin-5-yl)-phenyl]-ethyl}-2,3-dichloro-benzenesulfonamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H20Cl2N4O5S2 |
|---|
| Mol. Mass. | 579.475 |
|---|
| SMILES | Clc1cccc(c1Cl)S(=O)(=O)NC(Cc1ccc(cc1)C1CC(=O)NS1(=O)=O)c1nc2ccccc2[nH]1 |
|---|
| Structure | 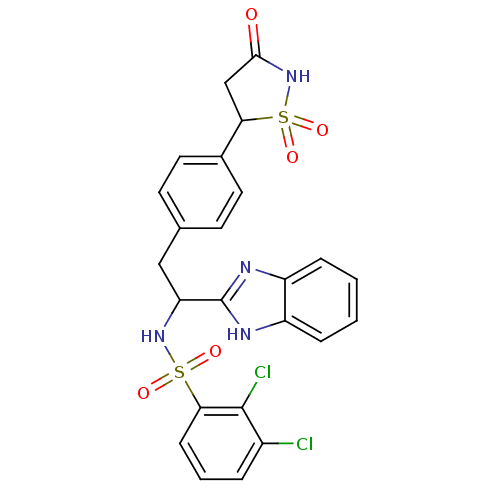
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Combs, AP; Zhu, W; Crawley, ML; Glass, B; Polam, P; Sparks, RB; Modi, D; Takvorian, A; McLaughlin, E; Yue, EW; Wasserman, Z; Bower, M; Wei, M; Rupar, M; Ala, PJ; Reid, BM; Ellis, D; Gonneville, L; Emm, T; Taylor, N; Yeleswaram, S; Li, Y; Wynn, R; Burn, TC; Hollis, G; Liu, PC; Metcalf, B Potent benzimidazole sulfonamide protein tyrosine phosphatase 1B inhibitors containing the heterocyclic (S)-isothiazolidinone phosphotyrosine mimetic. J Med Chem49:3774-89 (2006) [PubMed] Article
Combs, AP; Zhu, W; Crawley, ML; Glass, B; Polam, P; Sparks, RB; Modi, D; Takvorian, A; McLaughlin, E; Yue, EW; Wasserman, Z; Bower, M; Wei, M; Rupar, M; Ala, PJ; Reid, BM; Ellis, D; Gonneville, L; Emm, T; Taylor, N; Yeleswaram, S; Li, Y; Wynn, R; Burn, TC; Hollis, G; Liu, PC; Metcalf, B Potent benzimidazole sulfonamide protein tyrosine phosphatase 1B inhibitors containing the heterocyclic (S)-isothiazolidinone phosphotyrosine mimetic. J Med Chem49:3774-89 (2006) [PubMed] Article