| Citation |  Wishka, DG; Walker, DP; Yates, KM; Reitz, SC; Jia, S; Myers, JK; Olson, KL; Jacobsen, EJ; Wolfe, ML; Groppi, VE; Hanchar, AJ; Thornburgh, BA; Cortes-Burgos, LA; Wong, EH; Staton, BA; Raub, TJ; Higdon, NR; Wall, TM; Hurst, RS; Walters, RR; Hoffmann, WE; Hajos, M; Franklin, S; Carey, G; Gold, LH; Cook, KK; Sands, SB; Zhao, SX; Soglia, JR; Kalgutkar, AS; Arneric, SP; Rogers, BN Discovery of N-[(3R)-1-azabicyclo[2.2.2]oct-3-yl]furo[2,3-c]pyridine-5-carboxamide, an agonist of the alpha7 nicotinic acetylcholine receptor, for the potential treatment of cognitive deficits in schizophrenia: synthesis and structure--activity relationship. J Med Chem49:4425-36 (2006) [PubMed] Article Wishka, DG; Walker, DP; Yates, KM; Reitz, SC; Jia, S; Myers, JK; Olson, KL; Jacobsen, EJ; Wolfe, ML; Groppi, VE; Hanchar, AJ; Thornburgh, BA; Cortes-Burgos, LA; Wong, EH; Staton, BA; Raub, TJ; Higdon, NR; Wall, TM; Hurst, RS; Walters, RR; Hoffmann, WE; Hajos, M; Franklin, S; Carey, G; Gold, LH; Cook, KK; Sands, SB; Zhao, SX; Soglia, JR; Kalgutkar, AS; Arneric, SP; Rogers, BN Discovery of N-[(3R)-1-azabicyclo[2.2.2]oct-3-yl]furo[2,3-c]pyridine-5-carboxamide, an agonist of the alpha7 nicotinic acetylcholine receptor, for the potential treatment of cognitive deficits in schizophrenia: synthesis and structure--activity relationship. J Med Chem49:4425-36 (2006) [PubMed] Article |
|---|
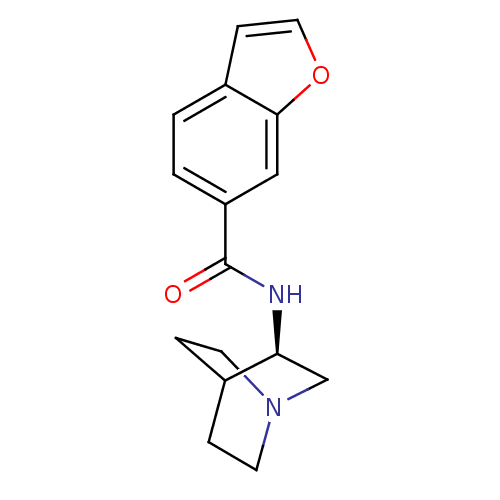
| SMILES | O=C(N[C@H]1CN2CCC1CC2)c1ccc2ccoc2c1 |wU:3.2,(27.6,-12.86,;27.61,-14.4,;28.95,-15.16,;30.28,-14.39,;30.27,-12.85,;31.61,-12.08,;32.95,-12.85,;32.94,-14.39,;31.62,-15.16,;30.74,-14.01,;31.56,-13.22,;26.28,-15.18,;26.29,-16.73,;24.94,-17.5,;23.61,-16.73,;22.15,-17.2,;21.24,-15.96,;22.15,-14.71,;23.61,-15.19,;24.94,-14.41,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wishka, DG; Walker, DP; Yates, KM; Reitz, SC; Jia, S; Myers, JK; Olson, KL; Jacobsen, EJ; Wolfe, ML; Groppi, VE; Hanchar, AJ; Thornburgh, BA; Cortes-Burgos, LA; Wong, EH; Staton, BA; Raub, TJ; Higdon, NR; Wall, TM; Hurst, RS; Walters, RR; Hoffmann, WE; Hajos, M; Franklin, S; Carey, G; Gold, LH; Cook, KK; Sands, SB; Zhao, SX; Soglia, JR; Kalgutkar, AS; Arneric, SP; Rogers, BN Discovery of N-[(3R)-1-azabicyclo[2.2.2]oct-3-yl]furo[2,3-c]pyridine-5-carboxamide, an agonist of the alpha7 nicotinic acetylcholine receptor, for the potential treatment of cognitive deficits in schizophrenia: synthesis and structure--activity relationship. J Med Chem49:4425-36 (2006) [PubMed] Article
Wishka, DG; Walker, DP; Yates, KM; Reitz, SC; Jia, S; Myers, JK; Olson, KL; Jacobsen, EJ; Wolfe, ML; Groppi, VE; Hanchar, AJ; Thornburgh, BA; Cortes-Burgos, LA; Wong, EH; Staton, BA; Raub, TJ; Higdon, NR; Wall, TM; Hurst, RS; Walters, RR; Hoffmann, WE; Hajos, M; Franklin, S; Carey, G; Gold, LH; Cook, KK; Sands, SB; Zhao, SX; Soglia, JR; Kalgutkar, AS; Arneric, SP; Rogers, BN Discovery of N-[(3R)-1-azabicyclo[2.2.2]oct-3-yl]furo[2,3-c]pyridine-5-carboxamide, an agonist of the alpha7 nicotinic acetylcholine receptor, for the potential treatment of cognitive deficits in schizophrenia: synthesis and structure--activity relationship. J Med Chem49:4425-36 (2006) [PubMed] Article