| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glycogen phosphorylase, liver form |
|---|
| Ligand | BDBM50194363 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_424351 (CHEMBL909554) |
|---|
| IC50 | >20000±n/a nM |
|---|
| Citation |  Li, YH; Coppo, FT; Evans, KA; Graybill, TL; Patel, M; Gale, J; Li, H; Tavares, F; Thomson, SA Synthesis and structure-activity relationships of 3-phenyl-2-propenamides as inhibitors of glycogen phosphorylase a. Bioorg Med Chem Lett16:5892-6 (2006) [PubMed] Article Li, YH; Coppo, FT; Evans, KA; Graybill, TL; Patel, M; Gale, J; Li, H; Tavares, F; Thomson, SA Synthesis and structure-activity relationships of 3-phenyl-2-propenamides as inhibitors of glycogen phosphorylase a. Bioorg Med Chem Lett16:5892-6 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glycogen phosphorylase, liver form |
|---|
| Name: | Glycogen phosphorylase, liver form |
|---|
| Synonyms: | Glycogen Phosphorylase (PYGL) | Glycogen Phosphorylase, liver form | Liver glycogen phosphorylase | PYGL | PYGL_HUMAN |
|---|
| Type: | Homodimer |
|---|
| Mol. Mass.: | 97153.98 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Dimers associate into a tetramer to form the enzymatically active phosphorylase A. |
|---|
| Residue: | 847 |
|---|
| Sequence: | MAKPLTDQEKRRQISIRGIVGVENVAELKKSFNRHLHFTLVKDRNVATTRDYYFALAHTV
RDHLVGRWIRTQQHYYDKCPKRVYYLSLEFYMGRTLQNTMINLGLQNACDEAIYQLGLDI
EELEEIEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEYGIFNQKIRDGWQVEEA
DDWLRYGNPWEKSRPEFMLPVHFYGKVEHTNTGTKWIDTQVVLALPYDTPVPGYMNNTVN
TMRLWSARAPNDFNLRDFNVGDYIQAVLDRNLAENISRVLYPNDNFFEGKELRLKQEYFV
VAATLQDIIRRFKASKFGSTRGAGTVFDAFPDQVAIQLNDTHPALAIPELMRIFVDIEKL
PWSKAWELTQKTFAYTNHTVLPEALERWPVDLVEKLLPRHLEIIYEINQKHLDRIVALFP
KDVDRLRRMSLIEEEGSKRINMAHLCIVGSHAVNGVAKIHSDIVKTKVFKDFSELEPDKF
QNKTNGITPRRWLLLCNPGLAELIAEKIGEDYVKDLSQLTKLHSFLGDDVFLRELAKVKQ
ENKLKFSQFLETEYKVKINPSSMFDVQVKRIHEYKRQLLNCLHVITMYNRIKKDPKKLFV
PRTVIIGGKAAPGYHMAKMIIKLITSVADVVNNDPMVGSKLKVIFLENYRVSLAEKVIPA
TDLSEQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENLFIFGMRIDDVA
ALDKKGYEAKEYYEALPELKLVIDQIDNGFFSPKQPDLFKDIINMLFYHDRFKVFADYEA
YVKCQDKVSQLYMNPKAWNTMVLKNIAASGKFSSDRTIKEYAQNIWNVEPSDLKISLSNE
SNKVNGN
|
|
|
|---|
| BDBM50194363 |
|---|
| n/a |
|---|
| Name | BDBM50194363 |
|---|
| Synonyms: | CHEMBL221442 | N-(3,4-dichlorophenyl)-4-(5-(3-(3,4-dichlorophenyl)acrylamido)pentyl)piperazine-1-carboxamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H28Cl4N4O2 |
|---|
| Mol. Mass. | 558.327 |
|---|
| SMILES | Clc1ccc(NC(=O)N2CCN(CCCCCNC(=O)\C=C\c3ccc(Cl)c(Cl)c3)CC2)cc1Cl |
|---|
| Structure | 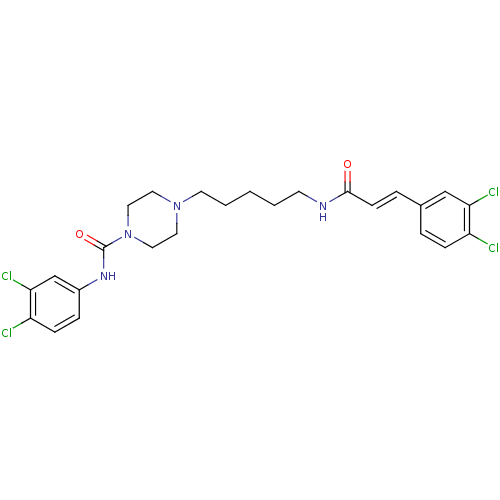
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, YH; Coppo, FT; Evans, KA; Graybill, TL; Patel, M; Gale, J; Li, H; Tavares, F; Thomson, SA Synthesis and structure-activity relationships of 3-phenyl-2-propenamides as inhibitors of glycogen phosphorylase a. Bioorg Med Chem Lett16:5892-6 (2006) [PubMed] Article
Li, YH; Coppo, FT; Evans, KA; Graybill, TL; Patel, M; Gale, J; Li, H; Tavares, F; Thomson, SA Synthesis and structure-activity relationships of 3-phenyl-2-propenamides as inhibitors of glycogen phosphorylase a. Bioorg Med Chem Lett16:5892-6 (2006) [PubMed] Article