

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Potassium voltage-gated channel subfamily H member 2 | ||
| Ligand | BDBM50609449 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2277957 | ||
| IC50 | 690±n/a nM | ||
| Citation |  Peng, X; Lanter, JC; Y-P Chen, A; Brand, MA; Wozniak, MK; Hoekman, S; Longin, O; Regeling, H; Zonneveld, W; P L Bell, R; Koenig, G; Hurst, RS; Blain, JF; Burnett, DA Discovery of oxazoline enhancers of cellular progranulin release. Bioorg Med Chem Lett80:0 (2023) [PubMed] Peng, X; Lanter, JC; Y-P Chen, A; Brand, MA; Wozniak, MK; Hoekman, S; Longin, O; Regeling, H; Zonneveld, W; P L Bell, R; Koenig, G; Hurst, RS; Blain, JF; Burnett, DA Discovery of oxazoline enhancers of cellular progranulin release. Bioorg Med Chem Lett80:0 (2023) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Potassium voltage-gated channel subfamily H member 2 | |||
| Name: | Potassium voltage-gated channel subfamily H member 2 | ||
| Synonyms: | 1,3-beta-glucan synthase component GLS2 | Cytochrome P450 3A4 | ERG | ERG1 | Eag-related protein 1 | Ether a-go-go related gene potassium channel (hERG) | Ether-a-go-go-related gene (HERG) | Ether-a-go-go-related gene potassium channel (hERG) | Ether-a-go-go-related gene potassium channel 1 | Ether-a-go-go-related gene potassium channel 1 (HERG) | Ether-a-go-go-related gene potassium channel 1 (hERG1) | Ether-a-go-go-related protein (hERG) | Ether-a-go-go-related protein 1 | Ether-a-go-go-related protein 1 (HERG) | H-ERG | HERG | KCNH2 | KCNH2_HUMAN | Potassium voltage-gated channel subfamily H member 2 (hERG) | Transcriptional regulator ERG | Voltage-gated potassium channel subunit Kv11.1 | eag homolog | hERG Potassium Channel 1 | putative potassium channel subunit | ||
| Type: | Multi-pass membrane protein | ||
| Mol. Mass.: | 126672.65 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q12809 | ||
| Residue: | 1159 | ||
| Sequence: |
| ||
| BDBM50609449 | |||
| n/a | |||
| Name | BDBM50609449 | ||
| Synonyms: | CHEMBL5290639 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H28FN3O | ||
| Mol. Mass. | 405.5077 | ||
| SMILES | Fc1ccc(cc1)[C@@H]1N(CCc2ccccc12)C1=NC[C@H](O1)C1CN2CCC1CC2 |r,wU:20.25,wD:7.7,t:20,(2.63,-4.63,;2.63,-3.09,;3.96,-2.33,;3.96,-.79,;2.63,-0,;1.3,-.79,;1.3,-2.33,;2.63,1.55,;1.28,2.32,;1.28,3.86,;2.63,4.63,;3.96,3.86,;5.3,4.63,;6.63,3.86,;6.63,2.32,;5.3,1.55,;3.96,2.32,;-.06,1.53,;-.05,-.01,;-1.54,-.5,;-2.43,.72,;-1.53,1.97,;-3.97,.72,;-5.39,1.37,;-6.63,.45,;-6.49,-1.1,;-5.07,-1.72,;-3.8,-.84,;-5.34,-.84,;-5.09,.44,)| | ||
| Structure | 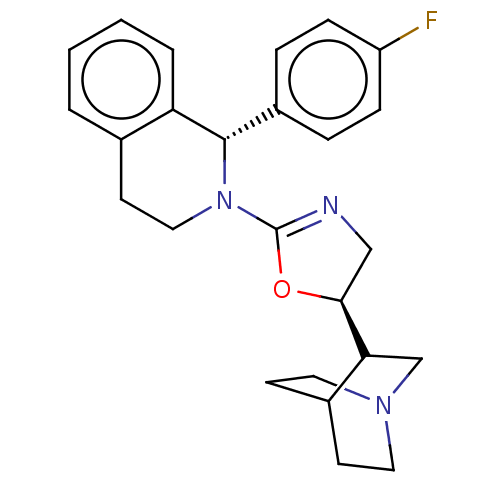
| ||