| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Ligand | BDBM50199954 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_422393 (CHEMBL909821) |
|---|
| IC50 | 9500±n/a nM |
|---|
| Citation |  Trapp, J; Jochum, A; Meier, R; Saunders, L; Marshall, B; Kunick, C; Verdin, E; Goekjian, P; Sippl, W; Jung, M Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem49:7307-16 (2006) [PubMed] Article Trapp, J; Jochum, A; Meier, R; Saunders, L; Marshall, B; Kunick, C; Verdin, E; Goekjian, P; Sippl, W; Jung, M Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem49:7307-16 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Synonyms: | NAD-Dependent Deacetylase Sirtuin-2 | NAD-dependent deacetylase sirtuin 2 | NAD-dependent deacetylase sirtuin 3 | NAD-dependent protein deacetylase sirtuin-2 (SIRT2) | SIR2-like | SIR2-like protein 2 | SIR2L | SIR2L2 | SIR2_HUMAN | SIRT2 | Sirtuin 2 (SIRT2) |
|---|
| Type: | Hydrolase |
|---|
| Mol. Mass.: | 43172.62 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 389 |
|---|
| Sequence: | MAEPDPSHPLETQAGKVQEAQDSDSDSEGGAAGGEADMDFLRNLFSQTLSLGSQKERLLD
ELTLEGVARYMQSERCRRVICLVGAGISTSAGIPDFRSPSTGLYDNLEKYHLPYPEAIFE
ISYFKKHPEPFFALAKELYPGQFKPTICHYFMRLLKDKGLLLRCYTQNIDTLERIAGLEQ
EDLVEAHGTFYTSHCVSASCRHEYPLSWMKEKIFSEVTPKCEDCQSLVKPDIVFFGESLP
ARFFSCMQSDFLKVDLLLVMGTSLQVQPFASLISKAPLSTPRLLINKEKAGQSDPFLGMI
MGLGGGMDFDSKKAYRDVAWLGECDQGCLALAELLGWKKELEDLVRREHASIDAQSGAGV
PNPSTSASPKKSPPPAKDEARTTEREKPQ
|
|
|
|---|
| BDBM50199954 |
|---|
| n/a |
|---|
| Name | BDBM50199954 |
|---|
| Synonyms: | 3-(4-fluoro-1H-indol-3-yl)-4-(1-(2-(1,2,5-trihydroxypentan-3-yloxy)ethyl)-1H-indol-3-yl)-1H-pyrrole-2,5-dione | CHEMBL218910 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H26FN3O6 |
|---|
| Mol. Mass. | 507.5102 |
|---|
| SMILES | OCCC(OCCn1cc(C2=C(C(=O)NC2=O)c2c[nH]c3cccc(F)c23)c2ccccc12)C(O)CO |t:10| |
|---|
| Structure | 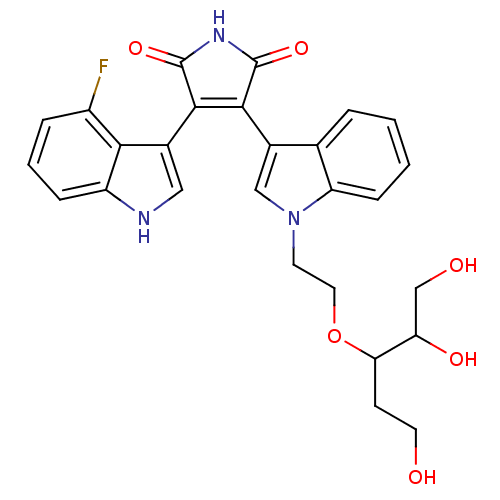
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Trapp, J; Jochum, A; Meier, R; Saunders, L; Marshall, B; Kunick, C; Verdin, E; Goekjian, P; Sippl, W; Jung, M Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem49:7307-16 (2006) [PubMed] Article
Trapp, J; Jochum, A; Meier, R; Saunders, L; Marshall, B; Kunick, C; Verdin, E; Goekjian, P; Sippl, W; Jung, M Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem49:7307-16 (2006) [PubMed] Article