| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Gamma-aminobutyric acid receptor subunit alpha-1/beta-2/gamma-2 |
|---|
| Ligand | BDBM50205615 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_435782 (CHEMBL904137) |
|---|
| Ki | 2.6±n/a nM |
|---|
| Citation |  Guerrini, G; Ciciani, G; Cambi, G; Bruni, F; Selleri, S; Besnard, F; Montali, M; Martini, C; Ghelardini, C; Galeotti, N; Costanzo, A Novel 3-iodo-8-ethoxypyrazolo[5,1-c][1,2,4]benzotriazine 5-oxide as promising lead for design of alpha5-inverse agonist useful tools for therapy of mnemonic damage. Bioorg Med Chem15:2573-86 (2007) [PubMed] Article Guerrini, G; Ciciani, G; Cambi, G; Bruni, F; Selleri, S; Besnard, F; Montali, M; Martini, C; Ghelardini, C; Galeotti, N; Costanzo, A Novel 3-iodo-8-ethoxypyrazolo[5,1-c][1,2,4]benzotriazine 5-oxide as promising lead for design of alpha5-inverse agonist useful tools for therapy of mnemonic damage. Bioorg Med Chem15:2573-86 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Gamma-aminobutyric acid receptor subunit alpha-1/beta-2/gamma-2 |
|---|
| Name: | Gamma-aminobutyric acid receptor subunit alpha-1/beta-2/gamma-2 |
|---|
| Synonyms: | GABA receptor alpha-1/beta-2/gamma-2 subunit | GABA-A receptor; alpha-1/beta-2/gamma-2 | GABAA Subunit α1β2γ2 | GABAA Subunit alpha-1, beta-2, gamma-2 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of EBI is 69455 |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Gamma-aminobutyric acid receptor subunit beta-2 |
|---|
| Synonyms: | GBRB2_RAT | Gabrb-2 | Gabrb2 | Gamma-aminobutyric acid receptor subunit beta-2 (GABA(A) beta-2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 54648.97 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P63138 |
|---|
| Residue: | 474 |
|---|
| Sequence: | MWRVRKRGYFGIWSFPLIIAAVCAQSVNDPSNMSLVKETVDRLLKGYDIRLRPDFGGPPV
AVGMNIDIASIDMVSEVNMDYTLTMYFQQAWRDKRLSYNVIPLNLTLDNRVADQLWVPDT
YFLNDKKSFVHGVTVKNRMIRLHPDGTVLYGLRITTTAACMMDLRRYPLDEQNCTLEIES
YGYTTDDIEFYWRGDDNAVTGVTKIELPQFSIVDYKLITKKVVFSTGSYPRLSLSFKLKR
NIGYFILQTYMPSILITILSWVSFWINYDASAARVALGITTVLTMTTINTHLRETLPKIP
YVKAIDMYLMGCFVFVFMALLEYALVNYIFFGRGPQRQKKAAEKAANANNEKMRLDVNKM
DPHENILLSTLEIKNEMATSEAVMGLGDPRSTMLAYDASSIQYRKAGLPRHSFGRNALER
HVAQKKSRLRRRASQLKITIPDLTDVNAIDRWSRIFFPVVFSFFNIVYWLYYVN
|
|
|
|---|
| Component 2 |
| Name: | Gamma-aminobutyric acid receptor subunit gamma-2 |
|---|
| Synonyms: | GBRG2_RAT | Gabrg2 | Gamma-aminobutyric acid receptor subunit gamma-2 (GABA(A) gamma-2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 54087.63 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P18508 |
|---|
| Residue: | 466 |
|---|
| Sequence: | MSSPNTWSTGSTVYSPVFSQKMTLWILLLLSLYPGFTSQKSDDDYEDYASNKTWVLTPKV
PEGDVTVILNNLLEGYDNKLRPDIGVKPTLIHTDMYVNSIGPVNAINMEYTIDIFFAQTW
YDRRLKFNSTIKVLRLNSNMVGKIWIPDTFFRNSKKADAHWITTPNRMLRIWNDGRVLYT
LRLTIDAECQLQLHNFPMDEHSCPLEFSSYGYPREEIVYQWKRSSVEVGDTRSWRLYQFS
FVGLRNTTEVVKTTSGDYVVMSVYFDLSRRMGYFTIQTYIPCTLIVVLSWVSFWINKDAV
PARTSLGITTVLTMTTLSTIARKSLPKVSYVTAMDLFVSVCFIFVFSALVEYGTLHYFVS
NRKPSKDKDKKKKNPAPTIDIRPRSATIQMNNATHLQERDEEYGYECLDGKDCASFFCCF
EDCRTGAWRHGRIHIRIAKMDSYARIFFPTAFCLFNLVYWVSYLYL
|
|
|
|---|
| Component 3 |
| Name: | Gamma-aminobutyric acid receptor subunit alpha-1 |
|---|
| Synonyms: | Benzodiazepine central | Benzodiazepine receptors | GABA A Benzodiazepine | GABA A Benzodiazepine Type I | GABA A Benzodiazepine Type II | GABA A Benzodiazepine Type IIL | GABA A Benzodiazepine Type IIM | GABA A Benzodiazepine omega1 | GABA A Benzodiazepine omega2 | GABA A Benzodiazepine omega5 | GABA A alpha1 | GABA A anti-Alpha1 | GABA receptor alpha-1 subunit | GABA, Chloride, TBOB | GABA-PICROTOXIN | GBRA1_RAT | Gabra-1 | Gabra1 | TBPS |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 51770.21 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P62813 |
|---|
| Residue: | 455 |
|---|
| Sequence: | MKKSRGLSDYLWAWTLILSTLSGRSYGQPSQDELKDNTTVFTRILDRLLDGYDNRLRPGL
GERVTEVKTDIFVTSFGPVSDHDMEYTIDVFFRQSWKDERLKFKGPMTVLRLNNLMASKI
WTPDTFFHNGKKSVAHNMTMPNKLLRITEDGTLLYTMRLTVRAECPMHLEDFPMDAHACP
LKFGSYAYTRAEVVYEWTREPARSVVVAEDGSRLNQYDLLGQTVDSGIVQSSTGEYVVMT
THFHLKRKIGYFVIQTYLPCIMTVILSQVSFWLNRESVPARTVFGVTTVLTMTTLSISAR
NSLPKVAYATAMDWFIAVCYAFVFSALIEFATVNYFTKRGYAWDGKSVVPEKPKKVKDPL
IKKNNTYAPTATSYTPNLARGDPGLATIAKSATIEPKEVKPETKPPEPKKTFNSVSKIDR
LSRIAFPLLFGIFNLVYWATYLNREPQLKAPTPHQ
|
|
|
|---|
| BDBM50205615 |
|---|
| n/a |
|---|
| Name | BDBM50205615 |
|---|
| Synonyms: | 3-iodo-8-ethoxypyrazolo[5,1-c][1,2,4]benzotriazine4-oxide | CHEMBL229145 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H9IN4O2 |
|---|
| Mol. Mass. | 356.1192 |
|---|
| SMILES | CCOc1ccc2n[n+]([O-])c3c(I)cnn3c2c1 |
|---|
| Structure | 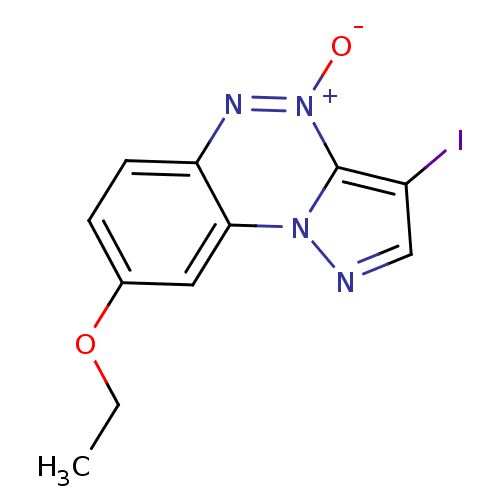
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Guerrini, G; Ciciani, G; Cambi, G; Bruni, F; Selleri, S; Besnard, F; Montali, M; Martini, C; Ghelardini, C; Galeotti, N; Costanzo, A Novel 3-iodo-8-ethoxypyrazolo[5,1-c][1,2,4]benzotriazine 5-oxide as promising lead for design of alpha5-inverse agonist useful tools for therapy of mnemonic damage. Bioorg Med Chem15:2573-86 (2007) [PubMed] Article
Guerrini, G; Ciciani, G; Cambi, G; Bruni, F; Selleri, S; Besnard, F; Montali, M; Martini, C; Ghelardini, C; Galeotti, N; Costanzo, A Novel 3-iodo-8-ethoxypyrazolo[5,1-c][1,2,4]benzotriazine 5-oxide as promising lead for design of alpha5-inverse agonist useful tools for therapy of mnemonic damage. Bioorg Med Chem15:2573-86 (2007) [PubMed] Article