| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sodium-dependent dopamine transporter |
|---|
| Ligand | BDBM50209971 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_458981 (CHEMBL925075) |
|---|
| Ki | 490±n/a nM |
|---|
| Citation |  Pruitt, JR; Batt, DG; Wacker, DA; Bostrom, LL; Booker, SK; McLaughlin, E; Houghton, GC; Varnes, JG; Christ, DD; Covington, M; Das, AM; Davies, P; Graden, D; Kariv, I; Orlovsky, Y; Stowell, NC; Vaddi, KG; Wadman, EA; Welch, PK; Yeleswaram, S; Solomon, KA; Newton, RC; Decicco, CP; Carter, PH; Ko, SS CC chemokine receptor-3 (CCR3) antagonists: improving the selectivity of DPC168 by reducing central ring lipophilicity. Bioorg Med Chem Lett17:2992-7 (2007) [PubMed] Article Pruitt, JR; Batt, DG; Wacker, DA; Bostrom, LL; Booker, SK; McLaughlin, E; Houghton, GC; Varnes, JG; Christ, DD; Covington, M; Das, AM; Davies, P; Graden, D; Kariv, I; Orlovsky, Y; Stowell, NC; Vaddi, KG; Wadman, EA; Welch, PK; Yeleswaram, S; Solomon, KA; Newton, RC; Decicco, CP; Carter, PH; Ko, SS CC chemokine receptor-3 (CCR3) antagonists: improving the selectivity of DPC168 by reducing central ring lipophilicity. Bioorg Med Chem Lett17:2992-7 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Sodium-dependent dopamine transporter |
|---|
| Name: | Sodium-dependent dopamine transporter |
|---|
| Synonyms: | DA transporter | DAT | DAT1 | Dopamine transporter (DAT) | Dopamine transporter protein (DAT) | Monoamine transporter | SC6A3_HUMAN | SLC6A3 | Sodium-dependent dopamine transporter | Sodium-dependent dopamine transporter (DAT) | Solute carrier family 6 member 3 |
|---|
| Type: | Multi-pass membrane protein |
|---|
| Mol. Mass.: | 68497.11 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q01959 |
|---|
| Residue: | 620 |
|---|
| Sequence: | MSKSKCSVGLMSSVVAPAKEPNAVGPKEVELILVKEQNGVQLTSSTLTNPRQSPVEAQDR
ETWGKKIDFLLSVIGFAVDLANVWRFPYLCYKNGGGAFLVPYLLFMVIAGMPLFYMELAL
GQFNREGAAGVWKICPILKGVGFTVILISLYVGFFYNVIIAWALHYLFSSFTTELPWIHC
NNSWNSPNCSDAHPGDSSGDSSGLNDTFGTTPAAEYFERGVLHLHQSHGIDDLGPPRWQL
TACLVLVIVLLYFSLWKGVKTSGKVVWITATMPYVVLTALLLRGVTLPGAIDGIRAYLSV
DFYRLCEASVWIDAATQVCFSLGVGFGVLIAFSSYNKFTNNCYRDAIVTTSINSLTSFSS
GFVVFSFLGYMAQKHSVPIGDVAKDGPGLIFIIYPEAIATLPLSSAWAVVFFIMLLTLGI
DSAMGGMESVITGLIDEFQLLHRHRELFTLFIVLATFLLSLFCVTNGGIYVFTLLDHFAA
GTSILFGVLIEAIGVAWFYGVGQFSDDIQQMTGQRPSLYWRLCWKLVSPCFLLFVVVVSI
VTFRPPHYGAYIFPDWANALGWVIATSSMAMVPIYAAYKFCSLPGSFREKLAYAIAPEKD
RELVDRGEVRQFTLRHWLKV
|
|
|
|---|
| BDBM50209971 |
|---|
| n/a |
|---|
| Name | BDBM50209971 |
|---|
| Synonyms: | 1-((1R,2S)-2-(((S)-3-(4-fluorobenzyl)piperidin-1-yl)methyl)cyclohexyl)-3-(3-acetylphenyl)urea | CHEMBL250689 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H36FN3O2 |
|---|
| Mol. Mass. | 465.6027 |
|---|
| SMILES | CC(=O)c1cccc(NC(=O)N[C@@H]2CCCC[C@H]2CN2CCC[C@@H](Cc3ccc(F)cc3)C2)c1 |
|---|
| Structure | 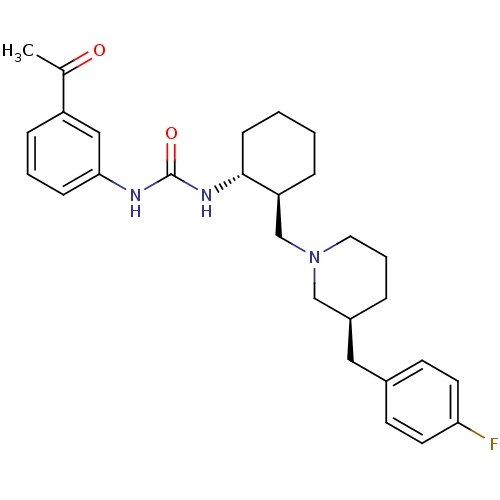
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pruitt, JR; Batt, DG; Wacker, DA; Bostrom, LL; Booker, SK; McLaughlin, E; Houghton, GC; Varnes, JG; Christ, DD; Covington, M; Das, AM; Davies, P; Graden, D; Kariv, I; Orlovsky, Y; Stowell, NC; Vaddi, KG; Wadman, EA; Welch, PK; Yeleswaram, S; Solomon, KA; Newton, RC; Decicco, CP; Carter, PH; Ko, SS CC chemokine receptor-3 (CCR3) antagonists: improving the selectivity of DPC168 by reducing central ring lipophilicity. Bioorg Med Chem Lett17:2992-7 (2007) [PubMed] Article
Pruitt, JR; Batt, DG; Wacker, DA; Bostrom, LL; Booker, SK; McLaughlin, E; Houghton, GC; Varnes, JG; Christ, DD; Covington, M; Das, AM; Davies, P; Graden, D; Kariv, I; Orlovsky, Y; Stowell, NC; Vaddi, KG; Wadman, EA; Welch, PK; Yeleswaram, S; Solomon, KA; Newton, RC; Decicco, CP; Carter, PH; Ko, SS CC chemokine receptor-3 (CCR3) antagonists: improving the selectivity of DPC168 by reducing central ring lipophilicity. Bioorg Med Chem Lett17:2992-7 (2007) [PubMed] Article