

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Vitamin D3 receptor | ||
| Ligand | BDBM50244268 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_491881 (CHEMBL946411) | ||
| EC50 | 0.017000±n/a nM | ||
| Citation |  Shimizu, M; Miyamoto, Y; Takaku, H; Matsuo, M; Nakabayashi, M; Masuno, H; Udagawa, N; DeLuca, HF; Ikura, T; Ito, N 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure. Bioorg Med Chem16:6949-64 (2008) [PubMed] Article Shimizu, M; Miyamoto, Y; Takaku, H; Matsuo, M; Nakabayashi, M; Masuno, H; Udagawa, N; DeLuca, HF; Ikura, T; Ito, N 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure. Bioorg Med Chem16:6949-64 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Vitamin D3 receptor | |||
| Name: | Vitamin D3 receptor | ||
| Synonyms: | 1,25-dihydroxyvitamin D3 receptor | Nr1i1 | Nuclear receptor subfamily 1 group I member 1 | VDR_MOUSE | Vdr | Vitamin D receptor | Vitamin D3 receptor | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 47830.18 | ||
| Organism: | Mus musculus | ||
| Description: | ChEMBL_491881 | ||
| Residue: | 422 | ||
| Sequence: |
| ||
| BDBM50244268 | |||
| n/a | |||
| Name | BDBM50244268 | ||
| Synonyms: | (20S)-1-alpha-2-alpha-25-Trihydroxy-2beta-methyl-16-ene-22-thia-26,27-dimethyl-19,24-dinorvitamin D3 | CHEMBL472153 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H44O4S | ||
| Mol. Mass. | 464.701 | ||
| SMILES | CCC(O)(CC)CS[C@@H](C)C1=CC[C@H]2\C(CCC[C@]12C)=C\C=C1C[C@@H](O)C(C)(O)[C@H](O)C1 |r,wU:13.12,18.20,24.26,wD:29.31,8.8,t:10,(-2.81,-9.71,;-3.92,-10.77,;-3.56,-12.27,;-2.08,-11.86,;-3.2,-13.77,;-1.72,-14.2,;-5.07,-12.59,;-6.1,-11.44,;-7.6,-11.76,;-8.63,-10.62,;-8.08,-13.23,;-7.18,-14.47,;-8.08,-15.71,;-9.54,-15.23,;-10.87,-16,;-12.22,-15.23,;-12.21,-13.69,;-10.87,-12.92,;-9.54,-13.7,;-9.55,-12.16,;-10.87,-17.54,;-12.22,-18.31,;-12.22,-19.85,;-10.88,-20.63,;-10.88,-22.17,;-9.54,-22.94,;-12.22,-22.93,;-11.12,-24.02,;-13.31,-24.02,;-13.53,-22.17,;-14.88,-22.94,;-13.53,-20.63,)| | ||
| Structure | 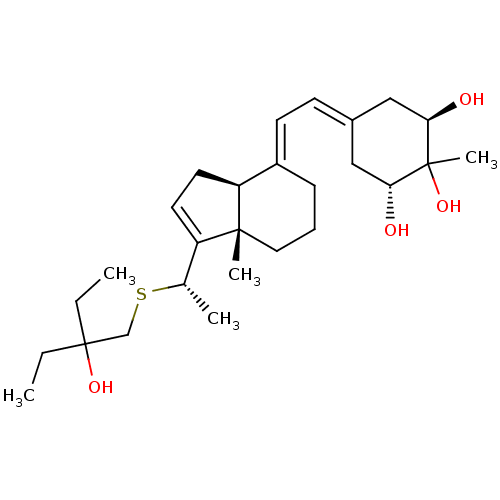
| ||