| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D(2) dopamine receptor |
|---|
| Ligand | BDBM50261729 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_514049 (CHEMBL979072) |
|---|
| EC50 | >50000±n/a nM |
|---|
| Citation |  DiMauro, EF; Buchanan, JL; Cheng, A; Emkey, R; Hitchcock, SA; Huang, L; Huang, MY; Janosky, B; Lee, JH; Li, X; Martin, MW; Tomlinson, SA; White, RD; Zheng, XM; Patel, VF; Fremeau, RT Structural modifications of N-arylamide oxadiazoles: Identification of N-arylpiperidine oxadiazoles as potent and selective agonists of CB2. Bioorg Med Chem Lett18:4267-74 (2008) [PubMed] Article DiMauro, EF; Buchanan, JL; Cheng, A; Emkey, R; Hitchcock, SA; Huang, L; Huang, MY; Janosky, B; Lee, JH; Li, X; Martin, MW; Tomlinson, SA; White, RD; Zheng, XM; Patel, VF; Fremeau, RT Structural modifications of N-arylamide oxadiazoles: Identification of N-arylpiperidine oxadiazoles as potent and selective agonists of CB2. Bioorg Med Chem Lett18:4267-74 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| D(2) dopamine receptor |
|---|
| Name: | D(2) dopamine receptor |
|---|
| Synonyms: | D(2) dopamine receptor | DOPAMINE D2 | DOPAMINE D2 Long | DOPAMINE D2 Short | DRD2 | DRD2_HUMAN | Dopamine D2 receptor | Dopamine D2 receptor (D2) | Dopamine D2 receptor (D2R) | Dopamine D2A | Dopamine2-like | d2 |
|---|
| Type: | Cell-surface receptors |
|---|
| Mol. Mass.: | 50647.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14416 |
|---|
| Residue: | 443 |
|---|
| Sequence: | MDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVS
REKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTA
SILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQN
ECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPL
KGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPP
SHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSR
RKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSA
VNPIIYTTFNIEFRKAFLKILHC
|
|
|
|---|
| BDBM50261729 |
|---|
| n/a |
|---|
| Name | BDBM50261729 |
|---|
| Synonyms: | 6-chloro-3-(4-(3-(2-chloro-4-fluorophenyl)-1,2,4-oxadiazol-5-yl)piperidin-1-yl)quinoline | CHEMBL468176 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H17Cl2FN4O |
|---|
| Mol. Mass. | 443.301 |
|---|
| SMILES | Fc1ccc(-c2noc(n2)C2CCN(CC2)c2cnc3ccc(Cl)cc3c2)c(Cl)c1 |
|---|
| Structure | 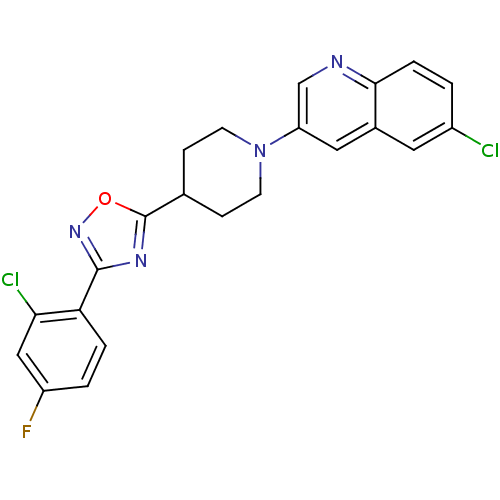
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 DiMauro, EF; Buchanan, JL; Cheng, A; Emkey, R; Hitchcock, SA; Huang, L; Huang, MY; Janosky, B; Lee, JH; Li, X; Martin, MW; Tomlinson, SA; White, RD; Zheng, XM; Patel, VF; Fremeau, RT Structural modifications of N-arylamide oxadiazoles: Identification of N-arylpiperidine oxadiazoles as potent and selective agonists of CB2. Bioorg Med Chem Lett18:4267-74 (2008) [PubMed] Article
DiMauro, EF; Buchanan, JL; Cheng, A; Emkey, R; Hitchcock, SA; Huang, L; Huang, MY; Janosky, B; Lee, JH; Li, X; Martin, MW; Tomlinson, SA; White, RD; Zheng, XM; Patel, VF; Fremeau, RT Structural modifications of N-arylamide oxadiazoles: Identification of N-arylpiperidine oxadiazoles as potent and selective agonists of CB2. Bioorg Med Chem Lett18:4267-74 (2008) [PubMed] Article