| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 2C9 |
|---|
| Ligand | BDBM50256667 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_515067 (CHEMBL1034809) |
|---|
| IC50 | 14000±n/a nM |
|---|
| Citation |  Sparks, SM; Banker, P; Bickett, DM; Clancy, DC; Dickerson, SH; Garrido, DM; Golden, PL; Peat, AJ; Sheckler, LR; Tavares, FX; Thomson, SA; Weiel, JE Anthranilimide-based glycogen phosphorylase inhibitors for the treatment of Type 2 diabetes: 2. Optimization of serine and threonine ether amino acid residues. Bioorg Med Chem Lett19:981-5 (2009) [PubMed] Article Sparks, SM; Banker, P; Bickett, DM; Clancy, DC; Dickerson, SH; Garrido, DM; Golden, PL; Peat, AJ; Sheckler, LR; Tavares, FX; Thomson, SA; Weiel, JE Anthranilimide-based glycogen phosphorylase inhibitors for the treatment of Type 2 diabetes: 2. Optimization of serine and threonine ether amino acid residues. Bioorg Med Chem Lett19:981-5 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 2C9 |
|---|
| Name: | Cytochrome P450 2C9 |
|---|
| Synonyms: | (R)-limonene 6-monooxygenase | (S)-limonene 6-monooxygenase | CP2C9_HUMAN | CYP2C10 | CYP2C9 | CYPIIC9 | Cytochrome P450 2C9 (CYP2C9 ) | Cytochrome P450 2C9 (CYP2C9) | P-450MP | P450 MP-4/MP-8 | P450 PB-1 | S-mephenytoin 4-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55636.33 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P11712 |
|---|
| Residue: | 490 |
|---|
| Sequence: | MDSLVVLVLCLSCLLLLSLWRQSSGRGKLPPGPTPLPVIGNILQIGIKDISKSLTNLSKV
YGPVFTLYFGLKPIVVLHGYEAVKEALIDLGEEFSGRGIFPLAERANRGFGIVFSNGKKW
KEIRRFSLMTLRNFGMGKRSIEDRVQEEARCLVEELRKTKASPCDPTFILGCAPCNVICS
IIFHKRFDYKDQQFLNLMEKLNENIKILSSPWIQICNNFSPIIDYFPGTHNKLLKNVAFM
KSYILEKVKEHQESMDMNNPQDFIDCFLMKMEKEKHNQPSEFTIESLENTAVDLFGAGTE
TTSTTLRYALLLLLKHPEVTAKVQEEIERVIGRNRSPCMQDRSHMPYTDAVVHEVQRYID
LLPTSLPHAVTCDIKFRNYLIPKGTTILISLTSVLHDNKEFPNPEMFDPHHFLDEGGNFK
KSKYFMPFSAGKRICVGEALAGMELFLFLTSILQNFNLKSLVDPKNLDTTPVVNGFASVP
PFYQLCFIPV
|
|
|
|---|
| BDBM50256667 |
|---|
| n/a |
|---|
| Name | BDBM50256667 |
|---|
| Synonyms: | (2S,3R)-3-isopropoxy-2-(3-(3-mesitylureido)-2-naphthamido)butanoic acid | CHEMBL473850 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H33N3O5 |
|---|
| Mol. Mass. | 491.5787 |
|---|
| SMILES | CC(C)O[C@H](C)[C@H](NC(=O)c1cc2ccccc2cc1NC(=O)Nc1c(C)cc(C)cc1C)C(O)=O |r| |
|---|
| Structure | 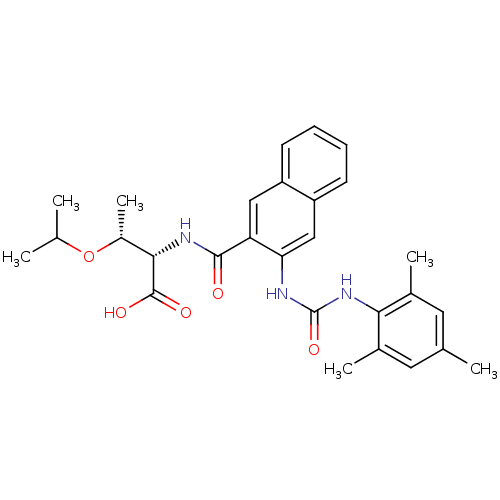
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sparks, SM; Banker, P; Bickett, DM; Clancy, DC; Dickerson, SH; Garrido, DM; Golden, PL; Peat, AJ; Sheckler, LR; Tavares, FX; Thomson, SA; Weiel, JE Anthranilimide-based glycogen phosphorylase inhibitors for the treatment of Type 2 diabetes: 2. Optimization of serine and threonine ether amino acid residues. Bioorg Med Chem Lett19:981-5 (2009) [PubMed] Article
Sparks, SM; Banker, P; Bickett, DM; Clancy, DC; Dickerson, SH; Garrido, DM; Golden, PL; Peat, AJ; Sheckler, LR; Tavares, FX; Thomson, SA; Weiel, JE Anthranilimide-based glycogen phosphorylase inhibitors for the treatment of Type 2 diabetes: 2. Optimization of serine and threonine ether amino acid residues. Bioorg Med Chem Lett19:981-5 (2009) [PubMed] Article