| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Ligand | BDBM50259057 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_500727 (CHEMBL967762) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Deninno, MP; Andrews, M; Bell, AS; Chen, Y; Eller-Zarbo, C; Eshelby, N; Etienne, JB; Moore, DE; Palmer, MJ; Visser, MS; Yu, LJ; Zavadoski, WJ; Michael Gibbs, E The discovery of potent, selective, and orally bioavailable PDE9 inhibitors as potential hypoglycemic agents. Bioorg Med Chem Lett19:2537-41 (2009) [PubMed] Article Deninno, MP; Andrews, M; Bell, AS; Chen, Y; Eller-Zarbo, C; Eshelby, N; Etienne, JB; Moore, DE; Palmer, MJ; Visser, MS; Yu, LJ; Zavadoski, WJ; Michael Gibbs, E The discovery of potent, selective, and orally bioavailable PDE9 inhibitors as potential hypoglycemic agents. Bioorg Med Chem Lett19:2537-41 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4B |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE4 | Isoform PDE4B1 | PDE32 | PDE4B | PDE4B1 | PDE4B_HUMAN | Phosphodiesterase 4B | Phosphodiesterase 4B (PDE4B) | Phosphodiesterase 4B (PDE4B1) | Phosphodiesterase Type 4 (PDE4B) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 83318.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07343 |
|---|
| Residue: | 736 |
|---|
| Sequence: | MKKSRSVMTVMADDNVKDYFECSLSKSYSSSSNTLGIDLWRGRRCCSGNLQLPPLSQRQS
ERARTPEGDGISRPTTLPLTTLPSIAITTVSQECFDVENGPSPGRSPLDPQASSSAGLVL
HATFPGHSQRRESFLYRSDSDYDLSPKAMSRNSSLPSEQHGDDLIVTPFAQVLASLRSVR
NNFTILTNLHGTSNKRSPAASQPPVSRVNPQEESYQKLAMETLEELDWCLDQLETIQTYR
SVSEMASNKFKRMLNRELTHLSEMSRSGNQVSEYISNTFLDKQNDVEIPSPTQKDREKKK
KQQLMTQISGVKKLMHSSSLNNTSISRFGVNTENEDHLAKELEDLNKWGLNIFNVAGYSH
NRPLTCIMYAIFQERDLLKTFRISSDTFITYMMTLEDHYHSDVAYHNSLHAADVAQSTHV
LLSTPALDAVFTDLEILAAIFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHL
AVGFKLLQEEHCDIFMNLTKKQRQTLRKMVIDMVLATDMSKHMSLLADLKTMVETKKVTS
SGVLLLDNYTDRIQVLRNMVHCADLSNPTKSLELYRQWTDRIMEEFFQQGDKERERGMEI
SPMCDKHTASVEKSQVGFIDYIVHPLWETWADLVQPDAQDILDTLEDNRNWYQSMIPQSP
SPPLDEQNRDCQGLMEKFQFELTLDEEDSEGPEKEGEGHSYFSSTKTLCVIDPENRDSLG
ETDIDIATEDKSPVDT
|
|
|
|---|
| BDBM50259057 |
|---|
| n/a |
|---|
| Name | BDBM50259057 |
|---|
| Synonyms: | 3-isopropyl-5-(2-(2-morpholinoethoxy)benzyl)-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-one | CHEMBL466068 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H27N5O3 |
|---|
| Mol. Mass. | 397.4708 |
|---|
| SMILES | CC(C)c1[nH]nc2c1nc(Cc1ccccc1OCCN1CCOCC1)[nH]c2=O |
|---|
| Structure | 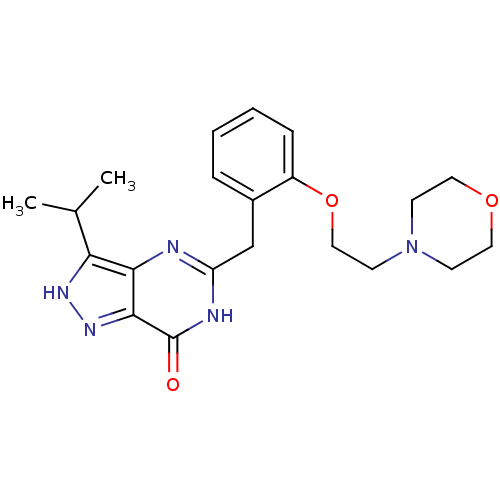
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Deninno, MP; Andrews, M; Bell, AS; Chen, Y; Eller-Zarbo, C; Eshelby, N; Etienne, JB; Moore, DE; Palmer, MJ; Visser, MS; Yu, LJ; Zavadoski, WJ; Michael Gibbs, E The discovery of potent, selective, and orally bioavailable PDE9 inhibitors as potential hypoglycemic agents. Bioorg Med Chem Lett19:2537-41 (2009) [PubMed] Article
Deninno, MP; Andrews, M; Bell, AS; Chen, Y; Eller-Zarbo, C; Eshelby, N; Etienne, JB; Moore, DE; Palmer, MJ; Visser, MS; Yu, LJ; Zavadoski, WJ; Michael Gibbs, E The discovery of potent, selective, and orally bioavailable PDE9 inhibitors as potential hypoglycemic agents. Bioorg Med Chem Lett19:2537-41 (2009) [PubMed] Article