| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glycogen phosphorylase, muscle form |
|---|
| Ligand | BDBM50277972 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_502059 (CHEMBL981270) |
|---|
| EC50 | >250000±n/a nM |
|---|
| Citation |  Dang, Q; Brown, BS; Liu, Y; Rydzewski, RM; Robinson, ED; van Poelje, PD; Reddy, MR; Erion, MD Fructose-1,6-bisphosphatase inhibitors. 1. Purine phosphonic acids as novel AMP mimics. J Med Chem52:2880-98 (2009) [PubMed] Article Dang, Q; Brown, BS; Liu, Y; Rydzewski, RM; Robinson, ED; van Poelje, PD; Reddy, MR; Erion, MD Fructose-1,6-bisphosphatase inhibitors. 1. Purine phosphonic acids as novel AMP mimics. J Med Chem52:2880-98 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glycogen phosphorylase, muscle form |
|---|
| Name: | Glycogen phosphorylase, muscle form |
|---|
| Synonyms: | Glycogen Phosphorylase (PYGM) | Glycogen phosphorylase a (RMGPa) | Glycogen phosphorylase, muscle form | Myophosphorylase | PYGM | PYGM_RABIT |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 97296.32 |
|---|
| Organism: | Oryctolagus cuniculus (rabbit) |
|---|
| Description: | Phosphorylation of Ser-15 converts phosphorylase B (unphosphorylated) to phosphorylase A. |
|---|
| Residue: | 843 |
|---|
| Sequence: | MSRPLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVKDRNVATPRDYYFALAHTV
RDHLVGRWIRTQQHYYEKDPKRIYYLSLEFYMGRTLQNTMVNLALENACDEATYQLGLDM
EELEEIEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEFGIFNQKICGGWQMEEA
DDWLRYGNPWEKARPEFTLPVHFYGRVEHTSQGAKWVDTQVVLAMPYDTPVPGYRNNVVN
TMRLWSAKAPNDFNLKDFNVGGYIQAVLDRNLAENISRVLYPNDNFFEGKELRLKQEYFV
VAATLQDIIRRFKSSKFGCRDPVRTNFDAFPDKVAIQLNDTHPSLAIPELMRVLVDLERL
DWDKAWEVTVKTCAYTNHTVLPEALERWPVHLLETLLPRHLQIIYEINQRFLNRVAAAFP
GDVDRLRRMSLVEEGAVKRINMAHLCIAGSHAVNGVARIHSEILKKTIFKDFYELEPHKF
QNKTNGITPRRWLVLCNPGLAEIIAERIGEEYISDLDQLRKLLSYVDDEAFIRDVAKVKQ
ENKLKFAAYLEREYKVHINPNSLFDVQVKRIHEYKRQLLNCLHVITLYNRIKKEPNKFVV
PRTVMIGGKAAPGYHMAKMIIKLITAIGDVVNHDPVVGDRLRVIFLENYRVSLAEKVIPA
ADLSEQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENFFIFGMRVEDVD
RLDQRGYNAQEYYDRIPELRQIIEQLSSGFFSPKQPDLFKDIVNMLMHHDRFKVFADYEE
YVKCQERVSALYKNPREWTRMVIRNIATSGKFSSDRTIAQYAREIWGVEPSRQRLPAPDE
KIP
|
|
|
|---|
| BDBM50277972 |
|---|
| n/a |
|---|
| Name | BDBM50277972 |
|---|
| Synonyms: | CHEMBL519503 | [5-(6-Amino-9-phenethyl-9H-purin-8-yl)-furan-2-yl]-phosphonic Acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H16N5O4P |
|---|
| Mol. Mass. | 385.3138 |
|---|
| SMILES | Nc1ncnc2n(CCc3ccccc3)c(nc12)-c1ccc(o1)P(O)(O)=O |
|---|
| Structure | 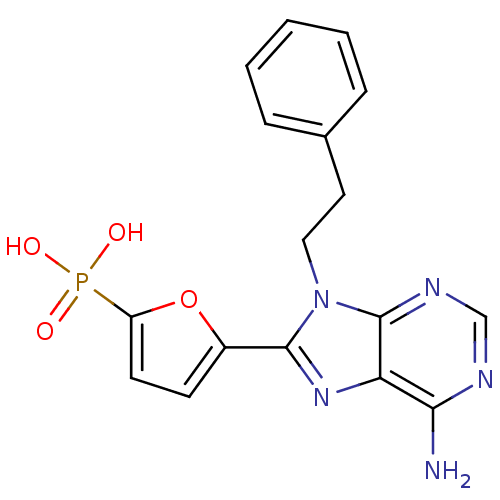
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Dang, Q; Brown, BS; Liu, Y; Rydzewski, RM; Robinson, ED; van Poelje, PD; Reddy, MR; Erion, MD Fructose-1,6-bisphosphatase inhibitors. 1. Purine phosphonic acids as novel AMP mimics. J Med Chem52:2880-98 (2009) [PubMed] Article
Dang, Q; Brown, BS; Liu, Y; Rydzewski, RM; Robinson, ED; van Poelje, PD; Reddy, MR; Erion, MD Fructose-1,6-bisphosphatase inhibitors. 1. Purine phosphonic acids as novel AMP mimics. J Med Chem52:2880-98 (2009) [PubMed] Article