| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Polyunsaturated fatty acid 5-lipoxygenase |
|---|
| Ligand | BDBM50012434 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_3964 (CHEMBL618062) |
|---|
| IC50 | 150±n/a nM |
|---|
| Citation |  Andrews, EG; Antognoli, GW; Breslow, R; Carta, MP; Carty, TJ; Chambers, RJ; Cheng, JB; Cohan, VL; Collins, JL; Damon, DB; Delehunt, J; Eggler, JF; Eskra, JD; Freiert, KW; Hada, WA; Marfat, A; Masamune, H; Melvin, LS; Mularski, CJ; Naclerio, BA Synthesis and pharmacological profile of two novel heterocyclic chromanols, CP-80,798 and CP-85,958, as potent LTD4 receptor antagonists Bioorg Med Chem Lett5:1365-1370 (1995) Article Andrews, EG; Antognoli, GW; Breslow, R; Carta, MP; Carty, TJ; Chambers, RJ; Cheng, JB; Cohan, VL; Collins, JL; Damon, DB; Delehunt, J; Eggler, JF; Eskra, JD; Freiert, KW; Hada, WA; Marfat, A; Masamune, H; Melvin, LS; Mularski, CJ; Naclerio, BA Synthesis and pharmacological profile of two novel heterocyclic chromanols, CP-80,798 and CP-85,958, as potent LTD4 receptor antagonists Bioorg Med Chem Lett5:1365-1370 (1995) Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Polyunsaturated fatty acid 5-lipoxygenase |
|---|
| Name: | Polyunsaturated fatty acid 5-lipoxygenase |
|---|
| Synonyms: | Alox5 | Arachidonate 5-lipoxygenase | LOX5_RAT |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 78082.31 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_1432947 |
|---|
| Residue: | 673 |
|---|
| Sequence: | MPSYTVTVATGSQWFAGTDDYIYLSLIGSAGCSEKHLLDKAFYNDFERGGRDSYDVTVDE
ELGEIYLVKIEKRKYRLHDDWYLKYITLKTPHDYIEFPCYRWITGEGEIVLRDGCAKLAR
DDQIHILKQHRRKELETRQKQYRWMEWNPGFPLSIDAKCHKDLPRDIQFDSEKGVDFVLN
YSKAMENLFINRFMHMFQSSWHDFADFEKIFVKISNTISERVKNHWQEDLMFGYQFLNGC
NPVLIKRCTELPKKLPVTTEMVECSLERQLSLEQEVQEGNIFIVDYELLDGIDANKTDPC
THQFLAAPICLLYKNLANKIVPIAIQLNQTPGEKNPIFLPTDSKYDWLLAKIWVRSSDFH
IHQTITHLLRTHLVSEVFGIAMYRQLPAVHPLFKLLVAHVRFTIAINTKAREQLNCEYGL
FDKANATGGGGHVQMVQRAVQDLTYSSLCFPEAIKARGMDNTEDIPYYFYRDDGLLVWEA
IQSFTTEVVSIYYEDDQVVEEDQELQDFVKDVYVYGMRGRKASGFPKSIKSREKLSEYLT
VVIFTASAQHAAVNFGQYDWCSWIPNAPPTMRAPPPTAKGVVTIEQIVDTLPDRGRSCWH
LGAVWALSQFQENELFLGMYPEEHFIEKPVKEAMIRFRKNLEAIVSVIAERNKNKKLPYY
YLSPDRIPNSVAI
|
|
|
|---|
| BDBM50012434 |
|---|
| n/a |
|---|
| Name | BDBM50012434 |
|---|
| Synonyms: | (REV-5,901)1-[3-(Quinolin-2-ylmethoxy)-phenyl]-hexan-1-ol | 1-[3-(Quinolin-2-ylmethoxy)-phenyl]-hexan-1-ol | 1-[3-(Quinolin-2-ylmethoxy)-phenyl]-hexan-1-ol (RG 5901) | 1-[3-(Quinolin-2-ylmethoxy)-phenyl]-hexan-1-ol(REV-5901) | CHEMBL8747 | REV-5901 | REV-901 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H25NO2 |
|---|
| Mol. Mass. | 335.4394 |
|---|
| SMILES | CCCCCC(O)c1cccc(OCc2ccc3ccccc3n2)c1 |
|---|
| Structure | 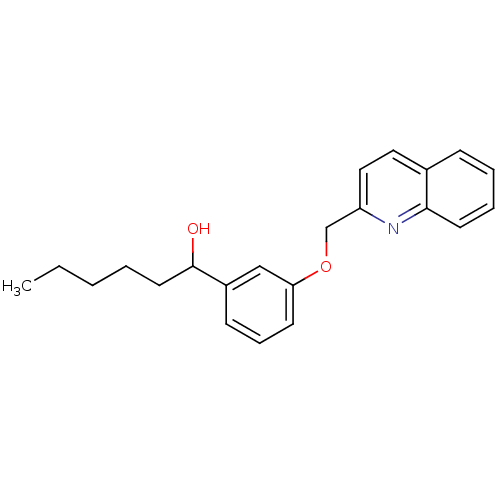
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Andrews, EG; Antognoli, GW; Breslow, R; Carta, MP; Carty, TJ; Chambers, RJ; Cheng, JB; Cohan, VL; Collins, JL; Damon, DB; Delehunt, J; Eggler, JF; Eskra, JD; Freiert, KW; Hada, WA; Marfat, A; Masamune, H; Melvin, LS; Mularski, CJ; Naclerio, BA Synthesis and pharmacological profile of two novel heterocyclic chromanols, CP-80,798 and CP-85,958, as potent LTD4 receptor antagonists Bioorg Med Chem Lett5:1365-1370 (1995) Article
Andrews, EG; Antognoli, GW; Breslow, R; Carta, MP; Carty, TJ; Chambers, RJ; Cheng, JB; Cohan, VL; Collins, JL; Damon, DB; Delehunt, J; Eggler, JF; Eskra, JD; Freiert, KW; Hada, WA; Marfat, A; Masamune, H; Melvin, LS; Mularski, CJ; Naclerio, BA Synthesis and pharmacological profile of two novel heterocyclic chromanols, CP-80,798 and CP-85,958, as potent LTD4 receptor antagonists Bioorg Med Chem Lett5:1365-1370 (1995) Article