| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytosolic phospholipase A2 |
|---|
| Ligand | BDBM50226800 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_493930 (CHEMBL947342) |
|---|
| IC50 | 100±n/a nM |
|---|
| Citation |  Chen, L; Wang, W; Lee, KL; Shen, MW; Murphy, EA; Zhang, W; Xu, X; Tam, S; Nickerson-Nutter, C; Goodwin, DG; Clark, JD; McKew, JC Reactions of functionalized sulfonamides: application to lowering the lipophilicity of cytosolic phospholipase A2alpha inhibitors. J Med Chem52:1156-71 (2009) [PubMed] Article Chen, L; Wang, W; Lee, KL; Shen, MW; Murphy, EA; Zhang, W; Xu, X; Tam, S; Nickerson-Nutter, C; Goodwin, DG; Clark, JD; McKew, JC Reactions of functionalized sulfonamides: application to lowering the lipophilicity of cytosolic phospholipase A2alpha inhibitors. J Med Chem52:1156-71 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytosolic phospholipase A2 |
|---|
| Name: | Cytosolic phospholipase A2 |
|---|
| Synonyms: | CPLA2 | Lysophospholipase | PA24A_HUMAN | PLA2G4 | PLA2G4A | Phosphatidylcholine 2-acylhydrolase | Phospholipase A2 | Phospholipase A2 group IVA |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 85219.30 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P47712 |
|---|
| Residue: | 749 |
|---|
| Sequence: | MSFIDPYQHIIVEHQYSHKFTVVVLRATKVTKGAFGDMLDTPDPYVELFISTTPDSRKRT
RHFNNDINPVWNETFEFILDPNQENVLEITLMDANYVMDETLGTATFTVSSMKVGEKKEV
PFIFNQVTEMVLEMSLEVCSCPDLRFSMALCDQEKTFRQQRKEHIRESMKKLLGPKNSEG
LHSARDVPVVAILGSGGGFRAMVGFSGVMKALYESGILDCATYVAGLSGSTWYMSTLYSH
PDFPEKGPEEINEELMKNVSHNPLLLLTPQKVKRYVESLWKKKSSGQPVTFTDIFGMLIG
ETLIHNRMNTTLSSLKEKVNTAQCPLPLFTCLHVKPDVSELMFADWVEFSPYEIGMAKYG
TFMAPDLFGSKFFMGTVVKKYEENPLHFLMGVWGSAFSILFNRVLGVSGSQSRGSTMEEE
LENITTKHIVSNDSSDSDDESHEPKGTENEDAGSDYQSDNQASWIHRMIMALVSDSALFN
TREGRAGKVHNFMLGLNLNTSYPLSPLSDFATQDSFDDDELDAAVADPDEFERIYEPLDV
KSKKIHVVDSGLTFNLPYPLILRPQRGVDLIISFDFSARPSDSSPPFKELLLAEKWAKMN
KLPFPKIDPYVFDREGLKECYVFKPKNPDMEKDCPTIIHFVLANINFRKYRAPGVPRETE
EEKEIADFDIFDDPESPFSTFNFQYPNQAFKRLHDLMHFNTLNNIDVIKEAMVESIEYRR
QNPSRCSVSLSNVEARRFFNKEFLSKPKA
|
|
|
|---|
| BDBM50226800 |
|---|
| n/a |
|---|
| Name | BDBM50226800 |
|---|
| Synonyms: | 4-(3-(1-benzhydryl-5-chloro-2-(2-(2-methoxyphenylsulfonamido)ethyl)-1H-indol-3-yl)propyl)benzoic acid | 4-{3-[1-benzhydryl-5-chloro-2-(2-{[(2-methoxyphenyl)sulfonyl]amino}ethyl)-1H-indol-3-yl]propyl}benzoic acid | CHEMBL408261 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C40H37ClN2O5S |
|---|
| Mol. Mass. | 693.25 |
|---|
| SMILES | COc1ccccc1S(=O)(=O)NCCc1c(CCCc2ccc(cc2)C(O)=O)c2cc(Cl)ccc2n1C(c1ccccc1)c1ccccc1 |
|---|
| Structure | 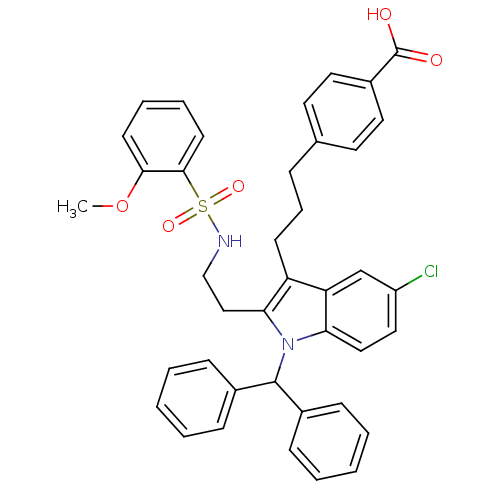
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chen, L; Wang, W; Lee, KL; Shen, MW; Murphy, EA; Zhang, W; Xu, X; Tam, S; Nickerson-Nutter, C; Goodwin, DG; Clark, JD; McKew, JC Reactions of functionalized sulfonamides: application to lowering the lipophilicity of cytosolic phospholipase A2alpha inhibitors. J Med Chem52:1156-71 (2009) [PubMed] Article
Chen, L; Wang, W; Lee, KL; Shen, MW; Murphy, EA; Zhang, W; Xu, X; Tam, S; Nickerson-Nutter, C; Goodwin, DG; Clark, JD; McKew, JC Reactions of functionalized sulfonamides: application to lowering the lipophilicity of cytosolic phospholipase A2alpha inhibitors. J Med Chem52:1156-71 (2009) [PubMed] Article