| Reaction Details |
|---|
 | Report a problem with these data |
| Target | MAP kinase-activated protein kinase 2 |
|---|
| Ligand | BDBM50305002 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_606910 (CHEMBL1072653) |
|---|
| IC50 | 1400±n/a nM |
|---|
| Citation |  Harris, CM; Ericsson, AM; Argiriadi, MA; Barberis, C; Borhani, DW; Burchat, A; Calderwood, DJ; Cunha, GA; Dixon, RW; Frank, KE; Johnson, EF; Kamens, J; Kwak, S; Li, B; Mullen, KD; Perron, DC; Wang, L; Wishart, N; Wu, X; Zhang, X; Zmetra, TR; Talanian, RV 2,4-Diaminopyrimidine MK2 inhibitors. Part II: Structure-based inhibitor optimization. Bioorg Med Chem Lett20:334-7 (2010) [PubMed] Article Harris, CM; Ericsson, AM; Argiriadi, MA; Barberis, C; Borhani, DW; Burchat, A; Calderwood, DJ; Cunha, GA; Dixon, RW; Frank, KE; Johnson, EF; Kamens, J; Kwak, S; Li, B; Mullen, KD; Perron, DC; Wang, L; Wishart, N; Wu, X; Zhang, X; Zmetra, TR; Talanian, RV 2,4-Diaminopyrimidine MK2 inhibitors. Part II: Structure-based inhibitor optimization. Bioorg Med Chem Lett20:334-7 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| MAP kinase-activated protein kinase 2 |
|---|
| Name: | MAP kinase-activated protein kinase 2 |
|---|
| Synonyms: | MAP kinase-activated protein kinase 2 (MAPKAPK2) | MAP kinase-activated protein kinase 2 (MK2) | MAP kinase-activated protein kinase 2 (p38/MK2) | MAPK-Activated Protein Kinase 2 (MK2) | MAPK-activated protein kinase 2 | MAPK2_HUMAN | MAPKAP kinase 2 | MAPKAPK-2 | MAPKAPK2 | MK2 |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 45579.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P49137 |
|---|
| Residue: | 400 |
|---|
| Sequence: | MLSNSQGQSPPVPFPAPAPPPQPPTPALPHPPAQPPPPPPQQFPQFHVKSGLQIKKNAII
DDYKVTSQVLGLGINGKVLQIFNKRTQEKFALKMLQDCPKARREVELHWRASQCPHIVRI
VDVYENLYAGRKCLLIVMECLDGGELFSRIQDRGDQAFTEREASEIMKSIGEAIQYLHSI
NIAHRDVKPENLLYTSKRPNAILKLTDFGFAKETTSHNSLTTPCYTPYYVAPEVLGPEKY
DKSCDMWSLGVIMYILLCGYPPFYSNHGLAISPGMKTRIRMGQYEFPNPEWSEVSEEVKM
LIRNLLKTEPTQRMTITEFMNHPWIMQSTKVPQTPLHTSRVLKEDKERWEDVKEEMTSAL
ATMRVDYEQIKIKKIEDASNPLLLKRRKKARALEAAALAH
|
|
|
|---|
| BDBM50305002 |
|---|
| n/a |
|---|
| Name | BDBM50305002 |
|---|
| Synonyms: | 7-(1H-indazol-5-yl)-5H-pyrrolo[3,2-d]pyrimidin-2-amine | CHEMBL591071 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H10N6 |
|---|
| Mol. Mass. | 250.2587 |
|---|
| SMILES | Nc1ncc2[nH]cc(-c3ccc4n[nH]cc4c3)c2n1 |
|---|
| Structure | 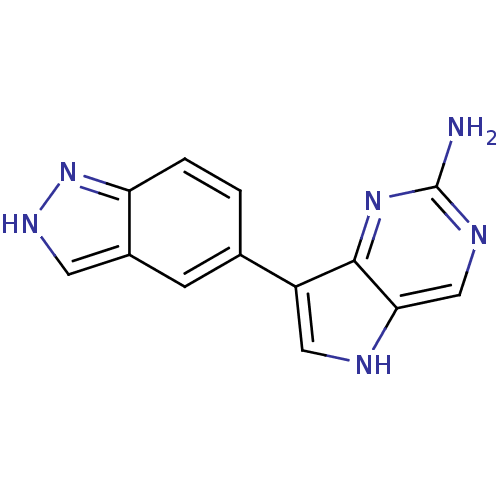
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Harris, CM; Ericsson, AM; Argiriadi, MA; Barberis, C; Borhani, DW; Burchat, A; Calderwood, DJ; Cunha, GA; Dixon, RW; Frank, KE; Johnson, EF; Kamens, J; Kwak, S; Li, B; Mullen, KD; Perron, DC; Wang, L; Wishart, N; Wu, X; Zhang, X; Zmetra, TR; Talanian, RV 2,4-Diaminopyrimidine MK2 inhibitors. Part II: Structure-based inhibitor optimization. Bioorg Med Chem Lett20:334-7 (2010) [PubMed] Article
Harris, CM; Ericsson, AM; Argiriadi, MA; Barberis, C; Borhani, DW; Burchat, A; Calderwood, DJ; Cunha, GA; Dixon, RW; Frank, KE; Johnson, EF; Kamens, J; Kwak, S; Li, B; Mullen, KD; Perron, DC; Wang, L; Wishart, N; Wu, X; Zhang, X; Zmetra, TR; Talanian, RV 2,4-Diaminopyrimidine MK2 inhibitors. Part II: Structure-based inhibitor optimization. Bioorg Med Chem Lett20:334-7 (2010) [PubMed] Article