| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM50317128 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_630653 (CHEMBL1111087) |
|---|
| EC50 | 66±n/a nM |
|---|
| Citation |  Ye, XY; Chen, S; Zhang, H; Locke, KT; O'Malley, K; Zhang, L; Srivastava, R; Miao, B; Meyers, D; Monshizadegan, H; Search, D; Grimm, D; Zhang, R; Lippy, J; Twamley, C; Muckelbauer, JK; Chang, C; An, Y; Hosagrahara, V; Zhang, L; Yang, TJ; Mukherjee, R; Cheng, PT; Tino, JA Synthesis and structure-activity relationships of 2-aryl-4-oxazolylmethoxy benzylglycines and 2-aryl-4-thiazolylmethoxy benzylglycines as novel, potent PPARalpha selective activators- PPARalpha and PPARgamma selectivity modulation. Bioorg Med Chem Lett20:2933-7 (2010) [PubMed] Article Ye, XY; Chen, S; Zhang, H; Locke, KT; O'Malley, K; Zhang, L; Srivastava, R; Miao, B; Meyers, D; Monshizadegan, H; Search, D; Grimm, D; Zhang, R; Lippy, J; Twamley, C; Muckelbauer, JK; Chang, C; An, Y; Hosagrahara, V; Zhang, L; Yang, TJ; Mukherjee, R; Cheng, PT; Tino, JA Synthesis and structure-activity relationships of 2-aryl-4-oxazolylmethoxy benzylglycines and 2-aryl-4-thiazolylmethoxy benzylglycines as novel, potent PPARalpha selective activators- PPARalpha and PPARgamma selectivity modulation. Bioorg Med Chem Lett20:2933-7 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM50317128 |
|---|
| n/a |
|---|
| Name | BDBM50317128 |
|---|
| Synonyms: | (S)-2-((1-(3-((2-(4-chlorophenyl)oxazol-4-yl)methoxy)phenyl)ethyl)((4-methoxyphenoxy)carbonyl)amino)acetic acid | CHEMBL1088114 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H25ClN2O7 |
|---|
| Mol. Mass. | 536.96 |
|---|
| SMILES | COc1ccc(OC(=O)N(CC(O)=O)[C@@H](C)c2cccc(OCc3coc(n3)-c3ccc(Cl)cc3)c2)cc1 |r| |
|---|
| Structure | 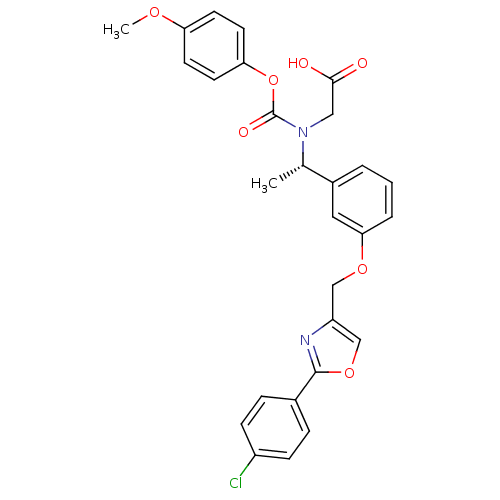
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ye, XY; Chen, S; Zhang, H; Locke, KT; O'Malley, K; Zhang, L; Srivastava, R; Miao, B; Meyers, D; Monshizadegan, H; Search, D; Grimm, D; Zhang, R; Lippy, J; Twamley, C; Muckelbauer, JK; Chang, C; An, Y; Hosagrahara, V; Zhang, L; Yang, TJ; Mukherjee, R; Cheng, PT; Tino, JA Synthesis and structure-activity relationships of 2-aryl-4-oxazolylmethoxy benzylglycines and 2-aryl-4-thiazolylmethoxy benzylglycines as novel, potent PPARalpha selective activators- PPARalpha and PPARgamma selectivity modulation. Bioorg Med Chem Lett20:2933-7 (2010) [PubMed] Article
Ye, XY; Chen, S; Zhang, H; Locke, KT; O'Malley, K; Zhang, L; Srivastava, R; Miao, B; Meyers, D; Monshizadegan, H; Search, D; Grimm, D; Zhang, R; Lippy, J; Twamley, C; Muckelbauer, JK; Chang, C; An, Y; Hosagrahara, V; Zhang, L; Yang, TJ; Mukherjee, R; Cheng, PT; Tino, JA Synthesis and structure-activity relationships of 2-aryl-4-oxazolylmethoxy benzylglycines and 2-aryl-4-thiazolylmethoxy benzylglycines as novel, potent PPARalpha selective activators- PPARalpha and PPARgamma selectivity modulation. Bioorg Med Chem Lett20:2933-7 (2010) [PubMed] Article