

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bile acid receptor | ||
| Ligand | BDBM50323541 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_644903 (CHEMBL1211233) | ||
| EC50 | 33±n/a nM | ||
| Citation |  Abel, U; Schlüter, T; Schulz, A; Hambruch, E; Steeneck, C; Hornberger, M; Hoffmann, T; Perovic-Ottstadt, S; Kinzel, O; Burnet, M; Deuschle, U; Kremoser, C Synthesis and pharmacological validation of a novel series of non-steroidal FXR agonists. Bioorg Med Chem Lett20:4911-7 (2010) [PubMed] Article Abel, U; Schlüter, T; Schulz, A; Hambruch, E; Steeneck, C; Hornberger, M; Hoffmann, T; Perovic-Ottstadt, S; Kinzel, O; Burnet, M; Deuschle, U; Kremoser, C Synthesis and pharmacological validation of a novel series of non-steroidal FXR agonists. Bioorg Med Chem Lett20:4911-7 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bile acid receptor | |||
| Name: | Bile acid receptor | ||
| Synonyms: | BAR | Bile acid receptor FXR | FXR | Farnesol receptor HRR-1 | HRR1 | NR1H4 | NR1H4_HUMAN | Nuclear receptor subfamily 1 group H member 4 | RIP14 | RXR-interacting protein 14 | Retinoid X receptor-interacting protein 14 | farnesoid x receptor | ||
| Type: | Nuclear Receptor | ||
| Mol. Mass.: | 55916.24 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q96RI1 | ||
| Residue: | 486 | ||
| Sequence: |
| ||
| BDBM50323541 | |||
| n/a | |||
| Name | BDBM50323541 | ||
| Synonyms: | 3-((6-((5-cyclopropyl-3-(2,6-dichlorophenyl)isoxazol-4-yl)methoxy)-2-(trifluoromethyl)pyridin-3-yl)methoxy)benzoic acid | CHEMBL1209293 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H19Cl2F3N2O5 | ||
| Mol. Mass. | 579.351 | ||
| SMILES | OC(=O)c1cccc(OCc2ccc(OCc3c(noc3C3CC3)-c3c(Cl)cccc3Cl)nc2C(F)(F)F)c1 |(-9.62,-34.92,;-8.29,-34.14,;-8.29,-32.6,;-6.96,-34.91,;-6.96,-36.46,;-5.62,-37.23,;-4.29,-36.46,;-4.29,-34.91,;-2.96,-34.13,;-1.62,-34.9,;-.29,-34.12,;-.31,-32.59,;1.02,-31.81,;2.36,-32.58,;3.7,-31.8,;5.04,-32.57,;6.36,-31.8,;7.62,-32.71,;8.87,-31.79,;8.39,-30.33,;6.83,-30.33,;5.92,-29.1,;5.75,-27.56,;4.51,-28.47,;7.63,-34.24,;8.96,-35,;10.29,-34.22,;8.97,-36.54,;7.64,-37.32,;6.3,-36.55,;6.29,-35.02,;4.96,-34.25,;2.37,-34.12,;1.04,-34.89,;1.04,-36.43,;-.29,-37.21,;2.38,-37.2,;1.03,-37.97,;-5.63,-34.14,)| | ||
| Structure | 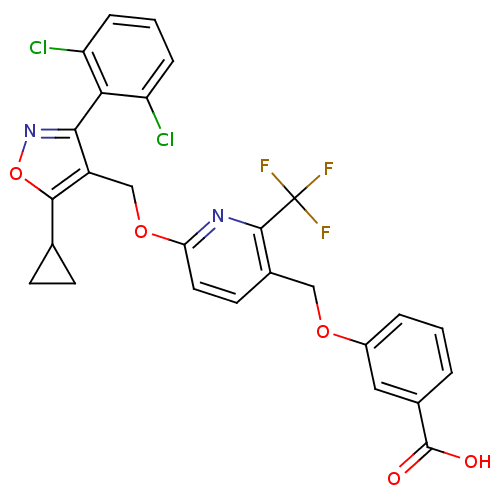
| ||