| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor 3 |
|---|
| Ligand | BDBM50094024 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_684662 (CHEMBL1285629) |
|---|
| EC50 | 1660±n/a nM |
|---|
| Citation |  Grove, SJ; Jamieson, C; Maclean, JK; Morrow, JA; Rankovic, Z Positive allosteric modulators of thea-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor. J Med Chem53:7271-9 (2010) [PubMed] Article Grove, SJ; Jamieson, C; Maclean, JK; Morrow, JA; Rankovic, Z Positive allosteric modulators of thea-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor. J Med Chem53:7271-9 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor 3 |
|---|
| Name: | Glutamate receptor 3 |
|---|
| Synonyms: | AMPA-selective glutamate receptor 3 | GLUR3 | GLURC | GRIA3 | GRIA3_HUMAN | GluR-3 | GluR-C | GluR-K3 | Glutamate receptor 3 | Glutamate receptor ionotropic AMPA | Glutamate receptor ionotropic, AMPA 3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 101172.14 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_468627 |
|---|
| Residue: | 894 |
|---|
| Sequence: | MARQKKMGQSVLRAVFFLVLGLLGHSHGGFPNTISIGGLFMRNTVQEHSAFRFAVQLYNT
NQNTTEKPFHLNYHVDHLDSSNSFSVTNAFCSQFSRGVYAIFGFYDQMSMNTLTSFCGAL
HTSFVTPSFPTDADVQFVIQMRPALKGAILSLLGHYKWEKFVYLYDTERGFSILQAIMEA
AVQNNWQVTARSVGNIKDVQEFRRIIEEMDRRQEKRYLIDCEVERINTILEQVVILGKHS
RGYHYMLANLGFTDILLERVMHGGANITGFQIVNNENPMVQQFIQRWVRLDEREFPEAKN
APLKYTSALTHDAILVIAEAFRYLRRQRVDVSRRGSAGDCLANPAVPWSQGIDIERALKM
VQVQGMTGNIQFDTYGRRTNYTIDVYEMKVSGSRKAGYWNEYERFVPFSDQQISNDSASS
ENRTIVVTTILESPYVMYKKNHEQLEGNERYEGYCVDLAYEIAKHVRIKYKLSIVGDGKY
GARDPETKIWNGMVGELVYGRADIAVAPLTITLVREEVIDFSKPFMSLGISIMIKKPQKS
KPGVFSFLDPLAYEIWMCIVFAYIGVSVVLFLVSRFSPYEWHLEDNNEEPRDPQSPPDPP
NEFGIFNSLWFSLGAFMQQGCDISPRSLSGRIVGGVWWFFTLIIISSYTANLAAFLTVER
MVSPIESAEDLAKQTEIAYGTLDSGSTKEFFRRSKIAVYEKMWSYMKSAEPSVFTKTTAD
GVARVRKSKGKFAFLLESTMNEYIEQRKPCDTMKVGGNLDSKGYGVATPKGSALRNAVNL
AVLKLNEQGLLDKLKNKWWYDKGECGSGGGDSKDKTSALSLSNVAGVFYILVGGLGLAMM
VALIEFCYKSRAESKRMKLTKNTQNFKPAPATNTQNYATYREGYNVYGTESVKI
|
|
|
|---|
| BDBM50094024 |
|---|
| n/a |
|---|
| Name | BDBM50094024 |
|---|
| Synonyms: | (+/-) Propane-2-sulfonic acid [2-(4'-cyano-biphenyl-4-yl)-propyl]-amide | CHEMBL435582 | propane-2-sulfonic acid [2-(4'-cyano-biphenyl-4-yl)-propyl]-amide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H22N2O2S |
|---|
| Mol. Mass. | 342.455 |
|---|
| SMILES | CC(C)S(=O)(=O)NCC(C)c1ccc(cc1)-c1ccc(cc1)C#N |
|---|
| Structure | 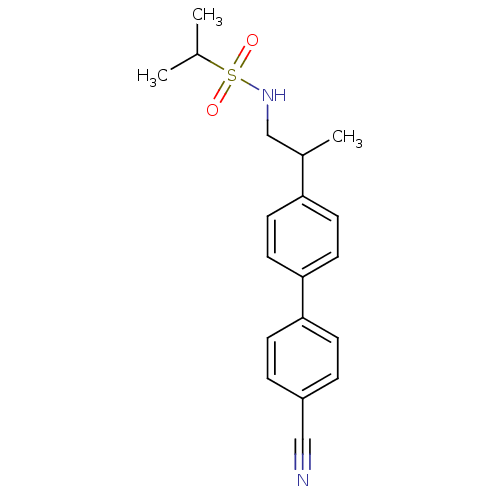
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Grove, SJ; Jamieson, C; Maclean, JK; Morrow, JA; Rankovic, Z Positive allosteric modulators of thea-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor. J Med Chem53:7271-9 (2010) [PubMed] Article
Grove, SJ; Jamieson, C; Maclean, JK; Morrow, JA; Rankovic, Z Positive allosteric modulators of thea-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor. J Med Chem53:7271-9 (2010) [PubMed] Article