| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Calcium/calmodulin-dependent protein kinase type II subunit beta |
|---|
| Ligand | BDBM15138 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_774307 (CHEMBL1908524) |
|---|
| Kd | 970±n/a nM |
|---|
| Citation |  Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Calcium/calmodulin-dependent protein kinase type II subunit beta |
|---|
| Name: | Calcium/calmodulin-dependent protein kinase type II subunit beta |
|---|
| Synonyms: | CAM2 | CAMK2 | CAMK2B | CAMKB | CaM kinase II | CaM kinase II beta | CaM kinase II subunit beta | CaM-kinase II beta chain | CaMK-II subunit beta | Calcium/calmodulin-dependent protein kinase type II | Calcium/calmodulin-dependent protein kinase type II beta chain | KCC2B_HUMAN |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 72685.48 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_26051204 |
|---|
| Residue: | 666 |
|---|
| Sequence: | MATTVTCTRFTDEYQLYEDIGKGAFSVVRRCVKLCTGHEYAAKIINTKKLSARDHQKLER
EARICRLLKHSNIVRLHDSISEEGFHYLVFDLVTGGELFEDIVAREYYSEADASHCIQQI
LEAVLHCHQMGVVHRDLKPENLLLASKCKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGY
LSPEVLRKEAYGKPVDIWACGVILYILLVGYPPFWDEDQHKLYQQIKAGAYDFPSPEWDT
VTPEAKNLINQMLTINPAKRITAHEALKHPWVCQRSTVASMMHRQETVECLKKFNARRKL
KGAILTTMLATRNFSVGRQTTAPATMSTAASGTTMGLVEQAKSLLNKKADGVKPQTNSTK
NSAAATSPKGTLPPAALEPQTTVIHNPVDGIKESSDSANTTIEDEDAKAPRVPDILSSVR
RGSGAPEAEGPLPCPSPAPFSPLPAPSPRISDILNSVRRGSGTPEAEGPLSAGPPPCLSP
ALLGPLSSPSPRISDILNSVRRGSGTPEAEGPSPVGPPPCPSPTIPGPLPTPSRKQEIIK
TTEQLIEAVNNGDFEAYAKICDPGLTSFEPEALGNLVEGMDFHRFYFENLLAKNSKPIHT
TILNPHVHVIGEDAACIAYIRLTQYIDGQGRPRTSQSEETRVWHRRDGKWQNVHFHCSGA
PVAPLQ
|
|
|
|---|
| BDBM15138 |
|---|
| n/a |
|---|
| Name | BDBM15138 |
|---|
| Synonyms: | 5-indazolyl pyridine 11g | 5-{5-[(2S)-2-amino-3-phenylpropoxy]pyridin-3-yl}-3-methyl-1H-indazole | A-674563 | Biochemistry 469551 Compound 11 | indazole-pyridine, 7 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H22N4O |
|---|
| Mol. Mass. | 358.4363 |
|---|
| SMILES | Cc1n[nH]c2ccc(cc12)-c1cncc(OC[C@@H](N)Cc2ccccc2)c1 |r| |
|---|
| Structure | 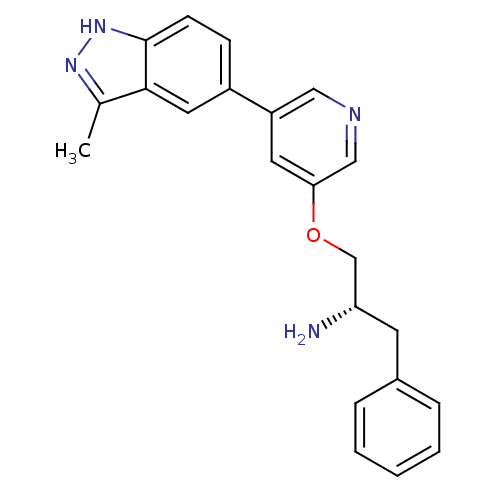
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article
Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article