| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 14 |
|---|
| Ligand | BDBM4552 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_774194 (CHEMBL1908411) |
|---|
| Kd | >10000±n/a nM |
|---|
| Citation |  Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 14 |
|---|
| Name: | Mitogen-activated protein kinase 14 |
|---|
| Synonyms: | CSAID-binding protein | CSBP | CSBP1 | CSBP2 | CSPB1 | Cytokine suppressive anti-inflammatory drug-binding protein | MAP kinase 14 | MAP kinase MXI2 | MAP kinase p38 alpha | MAPK 14 | MAPK14 | MAX-interacting protein 2 | MK14_HUMAN | MXI2 | Mitogen-activated protein kinase p38 alpha | SAPK2A | Stress-activated protein kinase 2a | p38 MAP kinase alpha/beta |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 41286.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q16539 |
|---|
| Residue: | 360 |
|---|
| Sequence: | MSQERPTFYRQELNKTIWEVPERYQNLSPVGSGAYGSVCAAFDTKTGLRVAVKKLSRPFQ
SIIHAKRTYRELRLLKHMKHENVIGLLDVFTPARSLEEFNDVYLVTHLMGADLNNIVKCQ
KLTDDHVQFLIYQILRGLKYIHSADIIHRDLKPSNLAVNEDCELKILDFGLARHTDDEMT
GYVATRWYRAPEIMLNWMHYNQTVDIWSVGCIMAELLTGRTLFPGTDHIDQLKLILRLVG
TPGAELLKKISSESARNYIQSLTQMPKMNFANVFIGANPLAVDLLEKMLVLDSDKRITAA
QALAHAYFAQYHDPDDEPVADPYDQSFESRDLLIDEWKSLTYDEVISFVPPPLDQEEMES
|
|
|
|---|
| BDBM4552 |
|---|
| n/a |
|---|
| Name | BDBM4552 |
|---|
| Synonyms: | 4-[(2,4-Dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methyl-1-piperazinyl)propoxy]-3-quinolinecarbonitrile | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile | BOSUTINIB | CHEMBL288441 | SKI-606 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H29Cl2N5O3 |
|---|
| Mol. Mass. | 530.446 |
|---|
| SMILES | COc1cc(Nc2c(cnc3cc(OCCCN4CCN(C)CC4)c(OC)cc23)C#N)c(Cl)cc1Cl |
|---|
| Structure | 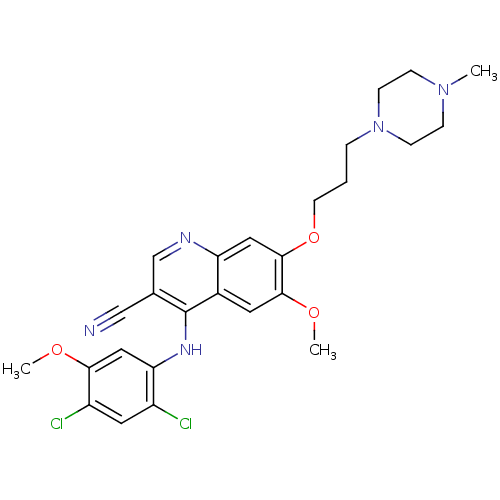
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article
Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article