| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 3 |
|---|
| Ligand | BDBM50161957 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_774365 (CHEMBL1908582) |
|---|
| Kd | >10000±n/a nM |
|---|
| Citation |  Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 3 |
|---|
| Name: | Mitogen-activated protein kinase 3 |
|---|
| Synonyms: | ERK1 | ERK1/ERK2 | Extracellular signal-regulated kinase 1 | Extracellular signal-regulated kinase 1 (ERK1) | MAP kinase ERK1 | MAPK3 | MAPK3/MAPK7 | MK03_HUMAN | Mitogen-activated protein kinase | Mitogen-activated protein kinase 3 | PRKM3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43136.58 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | curated by PDB 4QTB |
|---|
| Residue: | 379 |
|---|
| Sequence: | MAAAAAQGGGGGEPRRTEGVGPGVPGEVEMVKGQPFDVGPRYTQLQYIGEGAYGMVSSAY
DHVRKTRVAIKKISPFEHQTYCQRTLREIQILLRFRHENVIGIRDILRASTLEAMRDVYI
VQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLINTTCDL
KICDFGLARIADPEHDHTGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLS
NRPIFPGKHYLDQLNHILGILGSPSQEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSD
SKALDLLDRMLTFNPNKRITVEEALAHPYLEQYYDPTDEPVAEEPFTFAMELDDLPKERL
KELIFQETARFQPGVLEAP
|
|
|
|---|
| BDBM50161957 |
|---|
| n/a |
|---|
| Name | BDBM50161957 |
|---|
| Synonyms: | 4-Dimethylamino-but-2-enoic acid {4-[3-chloro-4-(pyridin-2-ylmethoxy)-phenylamino]-3-cyano-7-ethoxy-quinolin-6-yl}-amide | CHEMBL180022 | HKI-272 | N-(4-(3-chloro-4-(pyridin-2-ylmethoxy)phenylamino)-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)but-2-enamide | N-(4-(3-chloro-4-(pyridin-2-ylmethoxy)phenylamino)-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | NERATINIB | US10822334, Compound Neratinib | US11896597, Compound Neratinib | US20230382923, Compound Neratinib |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C30H29ClN6O3 |
|---|
| Mol. Mass. | 557.043 |
|---|
| SMILES | CCOc1cc2ncc(C#N)c(Nc3ccc(OCc4ccccn4)c(Cl)c3)c2cc1NC(=O)\C=C\CN(C)C |
|---|
| Structure | 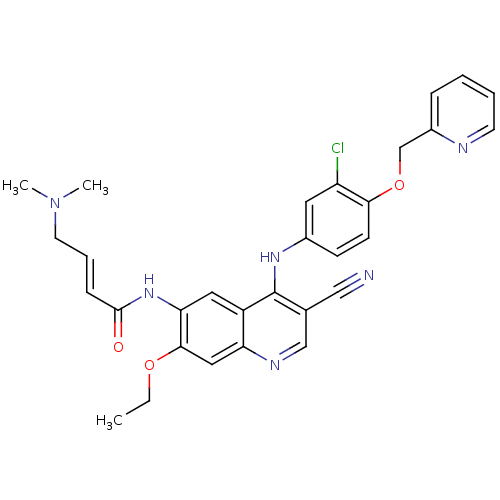
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article
Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article