| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha |
|---|
| Ligand | BDBM50357826 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_787242 (CHEMBL1918964) |
|---|
| IC50 | >1000±n/a nM |
|---|
| Citation |  Malamas, MS; Ni, Y; Erdei, J; Stange, H; Schindler, R; Lankau, HJ; Grunwald, C; Fan, KY; Parris, K; Langen, B; Egerland, U; Hage, T; Marquis, KL; Grauer, S; Brennan, J; Navarra, R; Graf, R; Harrison, BL; Robichaud, A; Kronbach, T; Pangalos, MN; Hoefgen, N; Brandon, NJ Highly potent, selective, and orally active phosphodiesterase 10A inhibitors. J Med Chem54:7621-38 (2011) [PubMed] Article Malamas, MS; Ni, Y; Erdei, J; Stange, H; Schindler, R; Lankau, HJ; Grunwald, C; Fan, KY; Parris, K; Langen, B; Egerland, U; Hage, T; Marquis, KL; Grauer, S; Brennan, J; Navarra, R; Graf, R; Harrison, BL; Robichaud, A; Kronbach, T; Pangalos, MN; Hoefgen, N; Brandon, NJ Highly potent, selective, and orally active phosphodiesterase 10A inhibitors. J Med Chem54:7621-38 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha |
|---|
| Name: | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha |
|---|
| Synonyms: | PDE6A | PDE6A_BOVIN | PDEA | Phosphodiesterase 6 | Phosphodiesterase 6A |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 99320.22 |
|---|
| Organism: | Bos taurus (Bovine) |
|---|
| Description: | P11541 |
|---|
| Residue: | 859 |
|---|
| Sequence: | MGEVTAEEVEKFLDSNVSFAKQYYNLRYRAKVISDLLGPREAAVDFSNYHALNSVEESEI
IFDLLRDFQDNLQAEKCVFNVMKKLCFLLQADRMSLFMYRARNGIAELATRLFNVHKDAV
LEECLVAPDSEIVFPLDMGVVGHVALSKKIVNVPNTEEDEHFCDFVDTLTEYQTKNILAS
PIMNGKDVVAIIMVVNKVDGPHFTENDEEILLKYLNFANLIMKVFHLSYLHNCETRRGQI
LLWSGSKVFEELTDIERQFHKALYTVRAFLNCDRYSVGLLDMTKQKEFFDVWPVLMGEAP
PYAGPRTPDGREINFYKVIDYILHGKEDIKVIPNPPPDHWALVSGLPTYVAQNGLICNIM
NAPSEDFFAFQKEPLDESGWMIKNVLSMPIVNKKEEIVGVATFYNRKDGKPFDEMDETLM
ESLTQFLGWSVLNPDTYELMNKLENRKDIFQDMVKYHVKCDNEEIQTILKTREVYGKEPW
ECEEEELAEILQGELPDADKYEINKFHFSDLPLTELELVKCGIQMYYELKVVDKFHIPQE
ALVRFMYSLSKGYRRITYHNWRHGFNVGQTMFSLLVTGKLKRYFTDLEALAMVTAAFCHD
IDHRGTNNLYQMKSQNPLAKLHGSSILERHHLEFGKTLLRDESLNIFQNLNRRQHEHAIH
MMDIAIIATDLALYFKKRTMFQKIVDQSKTYETQQEWTQYMMLDQTRKEIVMAMMMTACD
LSAITKPWEVQSKVALLVAAEFWEQGDLERTVLQQNPIPMMDRNKADELPKLQVGFIDFV
CTFVYKEFSRFHEEITPMLDGITNNRKEWKALADEYETKMKGLEEEKQKQQAANQAAAGS
QHGGKQPGGGPASKSCCVQ
|
|
|
|---|
| BDBM50357826 |
|---|
| n/a |
|---|
| Name | BDBM50357826 |
|---|
| Synonyms: | CHEMBL1915892 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H14ClFN4O |
|---|
| Mol. Mass. | 356.781 |
|---|
| SMILES | COc1ccc2nc(C)c3c(C)nc(-c4ccc(F)cc4Cl)n3c2n1 |
|---|
| Structure | 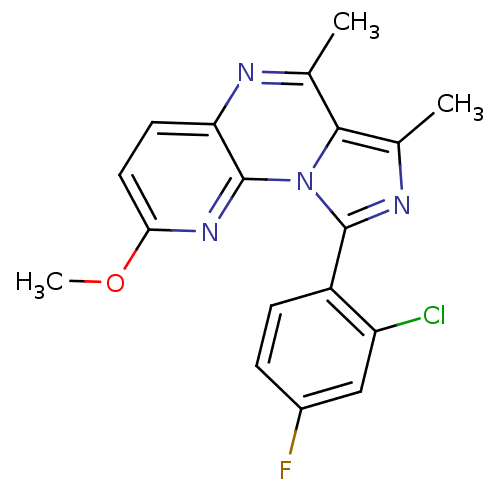
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Malamas, MS; Ni, Y; Erdei, J; Stange, H; Schindler, R; Lankau, HJ; Grunwald, C; Fan, KY; Parris, K; Langen, B; Egerland, U; Hage, T; Marquis, KL; Grauer, S; Brennan, J; Navarra, R; Graf, R; Harrison, BL; Robichaud, A; Kronbach, T; Pangalos, MN; Hoefgen, N; Brandon, NJ Highly potent, selective, and orally active phosphodiesterase 10A inhibitors. J Med Chem54:7621-38 (2011) [PubMed] Article
Malamas, MS; Ni, Y; Erdei, J; Stange, H; Schindler, R; Lankau, HJ; Grunwald, C; Fan, KY; Parris, K; Langen, B; Egerland, U; Hage, T; Marquis, KL; Grauer, S; Brennan, J; Navarra, R; Graf, R; Harrison, BL; Robichaud, A; Kronbach, T; Pangalos, MN; Hoefgen, N; Brandon, NJ Highly potent, selective, and orally active phosphodiesterase 10A inhibitors. J Med Chem54:7621-38 (2011) [PubMed] Article