| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 1A2 |
|---|
| Ligand | BDBM50358591 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_789255 (CHEMBL1924380) |
|---|
| IC50 | >40000±n/a nM |
|---|
| Citation |  Watterson, SH; Langevine, CM; Van Kirk, K; Kempson, J; Guo, J; Spergel, SH; Das, J; Moquin, RV; Dyckman, AJ; Nirschl, D; Gregor, K; Pattoli, MA; Yang, X; McIntyre, KW; Yang, G; Galella, MA; Booth-Lute, H; Chen, L; Yang, Z; Wang-Iverson, D; McKinnon, M; Dodd, JH; Barrish, JC; Burke, JR; Pitts, WJ Novel tricyclic inhibitors of IKK2: discovery and SAR leading to the identification of 2-methoxy-N-((6-(1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-7-yl)pyridin-2-yl)methyl)acetamide (BMS-066). Bioorg Med Chem Lett21:7006-12 (2011) [PubMed] Article Watterson, SH; Langevine, CM; Van Kirk, K; Kempson, J; Guo, J; Spergel, SH; Das, J; Moquin, RV; Dyckman, AJ; Nirschl, D; Gregor, K; Pattoli, MA; Yang, X; McIntyre, KW; Yang, G; Galella, MA; Booth-Lute, H; Chen, L; Yang, Z; Wang-Iverson, D; McKinnon, M; Dodd, JH; Barrish, JC; Burke, JR; Pitts, WJ Novel tricyclic inhibitors of IKK2: discovery and SAR leading to the identification of 2-methoxy-N-((6-(1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-7-yl)pyridin-2-yl)methyl)acetamide (BMS-066). Bioorg Med Chem Lett21:7006-12 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 1A2 |
|---|
| Name: | Cytochrome P450 1A2 |
|---|
| Synonyms: | CP1A2_HUMAN | CYP1A2 | CYPIA2 | Cholesterol 25-hydroxylase | Cytochrome P(3)450 | Cytochrome P450 1A | Cytochrome P450 1A2 (CYP1A2) | Cytochrome P450 4 | Cytochrome P450-P3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 58423.38 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P05177 |
|---|
| Residue: | 516 |
|---|
| Sequence: | MALSQSVPFSATELLLASAIFCLVFWVLKGLRPRVPKGLKSPPEPWGWPLLGHVLTLGKN
PHLALSRMSQRYGDVLQIRIGSTPVLVLSRLDTIRQALVRQGDDFKGRPDLYTSTLITDG
QSLTFSTDSGPVWAARRRLAQNALNTFSIASDPASSSSCYLEEHVSKEAKALISRLQELM
AGPGHFDPYNQVVVSVANVIGAMCFGQHFPESSDEMLSLVKNTHEFVETASSGNPLDFFP
ILRYLPNPALQRFKAFNQRFLWFLQKTVQEHYQDFDKNSVRDITGALFKHSKKGPRASGN
LIPQEKIVNLVNDIFGAGFDTVTTAISWSLMYLVTKPEIQRKIQKELDTVIGRERRPRLS
DRPQLPYLEAFILETFRHSSFLPFTIPHSTTRDTTLNGFYIPKKCCVFVNQWQVNHDPEL
WEDPSEFRPERFLTADGTAINKPLSEKMMLFGMGKRRCIGEVLAKWEIFLFLAILLQQLE
FSVPPGVKVDLTPIYGLTMKHARCEHVQARLRFSIN
|
|
|
|---|
| BDBM50358591 |
|---|
| n/a |
|---|
| Name | BDBM50358591 |
|---|
| Synonyms: | CHEMBL1923983 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H21N7O2 |
|---|
| Mol. Mass. | 379.4157 |
|---|
| SMILES | CNc1nc2[nH]c(cc2c2n(C)cnc12)-c1cccc(CNC(=O)COC)n1 |
|---|
| Structure | 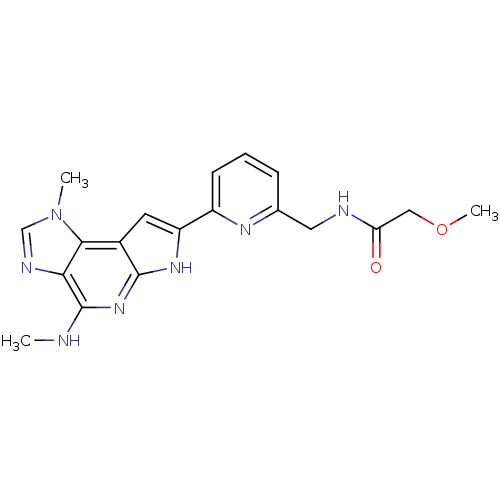
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Watterson, SH; Langevine, CM; Van Kirk, K; Kempson, J; Guo, J; Spergel, SH; Das, J; Moquin, RV; Dyckman, AJ; Nirschl, D; Gregor, K; Pattoli, MA; Yang, X; McIntyre, KW; Yang, G; Galella, MA; Booth-Lute, H; Chen, L; Yang, Z; Wang-Iverson, D; McKinnon, M; Dodd, JH; Barrish, JC; Burke, JR; Pitts, WJ Novel tricyclic inhibitors of IKK2: discovery and SAR leading to the identification of 2-methoxy-N-((6-(1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-7-yl)pyridin-2-yl)methyl)acetamide (BMS-066). Bioorg Med Chem Lett21:7006-12 (2011) [PubMed] Article
Watterson, SH; Langevine, CM; Van Kirk, K; Kempson, J; Guo, J; Spergel, SH; Das, J; Moquin, RV; Dyckman, AJ; Nirschl, D; Gregor, K; Pattoli, MA; Yang, X; McIntyre, KW; Yang, G; Galella, MA; Booth-Lute, H; Chen, L; Yang, Z; Wang-Iverson, D; McKinnon, M; Dodd, JH; Barrish, JC; Burke, JR; Pitts, WJ Novel tricyclic inhibitors of IKK2: discovery and SAR leading to the identification of 2-methoxy-N-((6-(1-methyl-4-(methylamino)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-7-yl)pyridin-2-yl)methyl)acetamide (BMS-066). Bioorg Med Chem Lett21:7006-12 (2011) [PubMed] Article