

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | MAP kinase-activated protein kinase 2 | ||
| Ligand | BDBM50362108 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_797472 (CHEMBL1943215) | ||
| IC50 | 4743±n/a nM | ||
| Citation |  Huang, X; Zhu, X; Chen, X; Zhou, W; Xiao, D; Degrado, S; Aslanian, R; Fossetta, J; Lundell, D; Tian, F; Trivedi, P; Palani, A A three-step protocol for lead optimization: quick identification of key conformational features and functional groups in the SAR studies of non-ATP competitive MK2 (MAPKAPK2) inhibitors. Bioorg Med Chem Lett22:65-70 (2011) [PubMed] Article Huang, X; Zhu, X; Chen, X; Zhou, W; Xiao, D; Degrado, S; Aslanian, R; Fossetta, J; Lundell, D; Tian, F; Trivedi, P; Palani, A A three-step protocol for lead optimization: quick identification of key conformational features and functional groups in the SAR studies of non-ATP competitive MK2 (MAPKAPK2) inhibitors. Bioorg Med Chem Lett22:65-70 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| MAP kinase-activated protein kinase 2 | |||
| Name: | MAP kinase-activated protein kinase 2 | ||
| Synonyms: | MAP kinase-activated protein kinase 2 (MAPKAPK2) | MAP kinase-activated protein kinase 2 (MK2) | MAP kinase-activated protein kinase 2 (p38/MK2) | MAPK-Activated Protein Kinase 2 (MK2) | MAPK-activated protein kinase 2 | MAPK2_HUMAN | MAPKAP kinase 2 | MAPKAPK-2 | MAPKAPK2 | MK2 | ||
| Type: | Serine/threonine-protein kinase | ||
| Mol. Mass.: | 45579.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P49137 | ||
| Residue: | 400 | ||
| Sequence: |
| ||
| BDBM50362108 | |||
| n/a | |||
| Name | BDBM50362108 | ||
| Synonyms: | CHEMBL1940510 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H31ClN4O2 | ||
| Mol. Mass. | 479.014 | ||
| SMILES | Clc1ccc(cc1)-c1ccc(o1)C(=O)N(Cc1ccccn1)C1CCC(CC1)N1CCNCC1 |(-.6,-7.34,;.94,-7.33,;1.7,-5.99,;3.24,-5.99,;4.01,-7.32,;3.25,-8.65,;1.72,-8.66,;5.55,-7.31,;6.47,-8.56,;7.93,-8.07,;7.93,-6.52,;6.46,-6.05,;9.26,-5.75,;9.25,-4.21,;10.59,-6.51,;10.6,-8.05,;11.94,-8.82,;11.93,-10.36,;13.27,-11.12,;14.6,-10.35,;14.59,-8.8,;13.25,-8.04,;11.92,-5.74,;13.26,-6.51,;14.59,-5.73,;14.58,-4.19,;13.24,-3.43,;11.91,-4.21,;15.91,-3.43,;17.25,-4.19,;18.57,-3.42,;18.57,-1.88,;17.24,-1.11,;15.9,-1.89,)| | ||
| Structure | 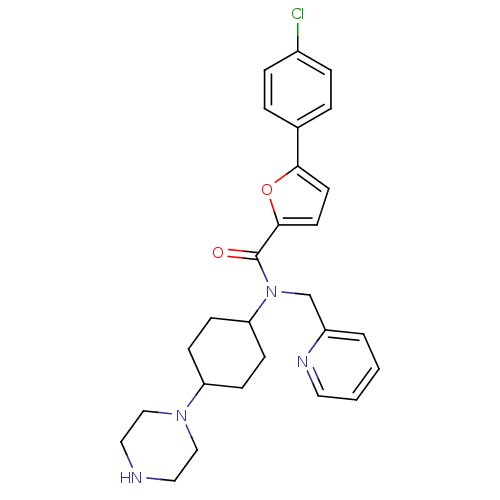
| ||