| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Matrix metalloproteinase-9 |
|---|
| Ligand | BDBM50106145 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_105530 |
|---|
| IC50 | 2±n/a nM |
|---|
| Citation |  Levin, JI; Chen, JM; Du, MT; Nelson, FC; Wehr, T; DiJoseph, JF; Killar, LM; Skala, S; Sung, A; Sharr, MA; Roth, CE; Jin, G; Cowling, R; Di, L; Sherman, M; Xu, ZB; March, CJ; Mohler, KM; Black, RA; Skotnicki, JS The discovery of anthranilic acid-based MMP inhibitors. Part 3: incorporation of basic amines. Bioorg Med Chem Lett11:2975-8 (2001) [PubMed] Levin, JI; Chen, JM; Du, MT; Nelson, FC; Wehr, T; DiJoseph, JF; Killar, LM; Skala, S; Sung, A; Sharr, MA; Roth, CE; Jin, G; Cowling, R; Di, L; Sherman, M; Xu, ZB; March, CJ; Mohler, KM; Black, RA; Skotnicki, JS The discovery of anthranilic acid-based MMP inhibitors. Part 3: incorporation of basic amines. Bioorg Med Chem Lett11:2975-8 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Matrix metalloproteinase-9 |
|---|
| Name: | Matrix metalloproteinase-9 |
|---|
| Synonyms: | 67 kDa matrix metalloproteinase-9 | 82 kDa matrix metalloproteinase-9 | 92 kDa gelatinase | 92 kDa type IV collagenase | CLG4B | GELB | Gelatinase B | MMP-9 | MMP9 | MMP9_HUMAN | Matrix metalloproteinase 9 (MMP-9) | Matrix metalloproteinase-9 (MMP-9) | Matrix metalloproteinase-9 (MMP9) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 78452.28 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14780 |
|---|
| Residue: | 707 |
|---|
| Sequence: | MSLWQPLVLVLLVLGCCFAAPRQRQSTLVLFPGDLRTNLTDRQLAEEYLYRYGYTRVAEM
RGESKSLGPALLLLQKQLSLPETGELDSATLKAMRTPRCGVPDLGRFQTFEGDLKWHHHN
ITYWIQNYSEDLPRAVIDDAFARAFALWSAVTPLTFTRVYSRDADIVIQFGVAEHGDGYP
FDGKDGLLAHAFPPGPGIQGDAHFDDDELWSLGKGVVVPTRFGNADGAACHFPFIFEGRS
YSACTTDGRSDGLPWCSTTANYDTDDRFGFCPSERLYTQDGNADGKPCQFPFIFQGQSYS
ACTTDGRSDGYRWCATTANYDRDKLFGFCPTRADSTVMGGNSAGELCVFPFTFLGKEYST
CTSEGRGDGRLWCATTSNFDSDKKWGFCPDQGYSLFLVAAHEFGHALGLDHSSVPEALMY
PMYRFTEGPPLHKDDVNGIRHLYGPRPEPEPRPPTTTTPQPTAPPTVCPTGPPTVHPSER
PTAGPTGPPSAGPTGPPTAGPSTATTVPLSPVDDACNVNIFDAIAEIGNQLYLFKDGKYW
RFSEGRGSRPQGPFLIADKWPALPRKLDSVFEERLSKKLFFFSGRQVWVYTGASVLGPRR
LDKLGLGADVAQVTGALRSGRGKMLLFSGRRLWRFDVKAQMVDPRSASEVDRMFPGVPLD
THDVFQYREKAYFCQDRFYWRVSSRSELNQVDQVGYVTYDILQCPED
|
|
|
|---|
| BDBM50106145 |
|---|
| n/a |
|---|
| Name | BDBM50106145 |
|---|
| Synonyms: | 5-Bromo-N-hydroxy-2-{(4-methoxy-benzenesulfonyl)-[4-(2-piperidin-1-yl-ethoxy)-benzyl]-amino}-3-methyl-benzamide | CHEMBL99745 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H34BrN3O6S |
|---|
| Mol. Mass. | 632.566 |
|---|
| SMILES | COc1ccc(cc1)S(=O)(=O)N(Cc1ccc(OCCN2CCCCC2)cc1)c1c(C)cc(Br)cc1C(=O)NO |
|---|
| Structure | 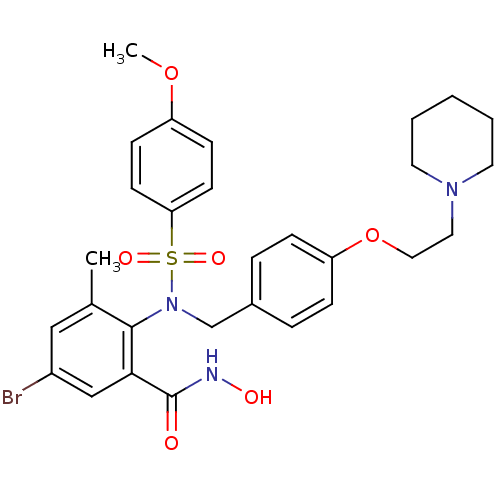
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Levin, JI; Chen, JM; Du, MT; Nelson, FC; Wehr, T; DiJoseph, JF; Killar, LM; Skala, S; Sung, A; Sharr, MA; Roth, CE; Jin, G; Cowling, R; Di, L; Sherman, M; Xu, ZB; March, CJ; Mohler, KM; Black, RA; Skotnicki, JS The discovery of anthranilic acid-based MMP inhibitors. Part 3: incorporation of basic amines. Bioorg Med Chem Lett11:2975-8 (2001) [PubMed]
Levin, JI; Chen, JM; Du, MT; Nelson, FC; Wehr, T; DiJoseph, JF; Killar, LM; Skala, S; Sung, A; Sharr, MA; Roth, CE; Jin, G; Cowling, R; Di, L; Sherman, M; Xu, ZB; March, CJ; Mohler, KM; Black, RA; Skotnicki, JS The discovery of anthranilic acid-based MMP inhibitors. Part 3: incorporation of basic amines. Bioorg Med Chem Lett11:2975-8 (2001) [PubMed]