

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Substance-K receptor | ||
| Ligand | BDBM50045039 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_141787 | ||
| IC50 | 244±n/a nM | ||
| Citation |  Kucharczyk, N; Thurieau, C; Paladino, J; Morris, AD; Bonnet, J; Canet, E; Krause, JE; Regoli, D; Couture, R; Fauchère, JL Tetrapeptide tachykinin antagonists: synthesis and modulation of the physicochemical and pharmacological properties of a new series of partially cyclic analogs. J Med Chem36:1654-61 (1993) [PubMed] Kucharczyk, N; Thurieau, C; Paladino, J; Morris, AD; Bonnet, J; Canet, E; Krause, JE; Regoli, D; Couture, R; Fauchère, JL Tetrapeptide tachykinin antagonists: synthesis and modulation of the physicochemical and pharmacological properties of a new series of partially cyclic analogs. J Med Chem36:1654-61 (1993) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Substance-K receptor | |||
| Name: | Substance-K receptor | ||
| Synonyms: | NK-2 receptor | NK-2R | NK2R_RAT | Neurokinin 2 receptor | Neurokinin A receptor | Neurokinin NK2 | SKR | Substance-K receptor | Tac2r | Tachykinin receptor 2 | Tacr2 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 43868.63 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | Neurokinin NK2 TACR2 Murine::P16610 | ||
| Residue: | 390 | ||
| Sequence: |
| ||
| BDBM50045039 | |||
| n/a | |||
| Name | BDBM50045039 | ||
| Synonyms: | 6-(3-{2-[1-(Benzyl-methyl-carbamoyl)-2-phenyl-ethylcarbamoyl]-2-[2-(3,6-dioxo-2,5-diaza-tricyclo[6.2.2.0*2,7*]dodec-4-yl)-acetylamino]-ethyl}-indole-1-carbonyloxy)-hexyl-ammonium | CHEMBL3144210 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C47H57N7O7 | ||
| Mol. Mass. | 831.9982 | ||
| SMILES | CN(Cc1ccccc1)C(=O)[C@H](Cc1ccccc1)NC(=O)[C@@H](Cc1cn(C(=O)OCCCCCCN)c2ccccc12)NC(=O)C[C@@H]1NC(=O)[C@@H]2C3CCC(CC3)N2C1=O |r,wU:51.54,47.50,22.24,wD:11.12,THB:49:51:54.53:56.57,(11.28,-9.71,;10.14,-8.66,;11.3,-7.64,;12.76,-8.14,;13.92,-7.12,;15.38,-7.61,;15.68,-9.13,;14.52,-10.14,;13.06,-9.65,;8.69,-8.17,;8.38,-6.66,;7.53,-9.18,;7.83,-10.69,;9.29,-11.19,;10.45,-10.17,;11.91,-10.66,;12.21,-12.17,;11.05,-13.19,;9.59,-12.7,;6.07,-8.69,;4.91,-9.71,;5.22,-11.22,;3.46,-9.21,;2.3,-10.23,;2.6,-11.74,;4,-12.38,;3.82,-13.91,;4.95,-14.96,;6.42,-14.5,;4.61,-16.46,;5.75,-17.5,;5.41,-19.01,;6.54,-20.05,;6.2,-21.55,;7.33,-22.6,;6.99,-24.1,;8.13,-25.14,;2.31,-14.22,;1.52,-15.54,;-.02,-15.52,;-.77,-14.18,;.02,-12.85,;1.56,-12.87,;3.15,-7.7,;4.31,-6.69,;5.77,-7.18,;4.01,-5.18,;2.55,-4.68,;2.25,-3.17,;.79,-2.68,;.49,-1.17,;-.37,-3.7,;-1.73,-3.16,;-3.23,-3.82,;-3.03,-5.23,;-1.46,-4.56,;-1.2,-2.62,;-1.66,-1.49,;-.07,-5.21,;1.39,-5.7,;1.69,-7.21,)| | ||
| Structure | 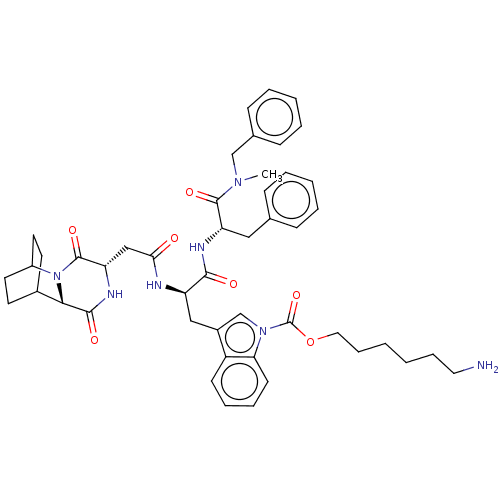
| ||