| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neprilysin |
|---|
| Ligand | BDBM50050158 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_144463 (CHEMBL755110) |
|---|
| IC50 | 1.3±n/a nM |
|---|
| Citation |  Coric, P; Turcaud, S; Meudal, H; Roques, BP; Fournie-Zaluski, MC Optimal recognition of neutral endopeptidase and angiotensin-converting enzyme active sites by mercaptoacyldipeptides as a means to design potent dual inhibitors. J Med Chem39:1210-9 (1996) [PubMed] Article Coric, P; Turcaud, S; Meudal, H; Roques, BP; Fournie-Zaluski, MC Optimal recognition of neutral endopeptidase and angiotensin-converting enzyme active sites by mercaptoacyldipeptides as a means to design potent dual inhibitors. J Med Chem39:1210-9 (1996) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Neprilysin |
|---|
| Name: | Neprilysin |
|---|
| Synonyms: | Atriopeptidase | CD10 antigen | CD_antigen=CD10 | Enkephalinase | MME | NEP | NEP_RABIT | Neutral Endopeptidase | Neutral endopeptidase 24.11 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 85570.75 |
|---|
| Organism: | Oryctolagus cuniculus (rabbit) |
|---|
| Description: | NEP was purified to homogeneity from rabbit kidney. |
|---|
| Residue: | 750 |
|---|
| Sequence: | MGRSESQMDITDINTPKPKKKQRWTPLEISLSVLVLLLTVIAVTMIALYATYDDGICKSS
DCIKSAARLIQNMDATAEPCTDFFKYACGGWLKRNVIPETSSRYSNFDILRDELEVILKD
VLQEPKTEDIVAVQKAKTLYRSCVNETAIDSRGGQPLLKLLPDVYGWPVATQNWEQTYGT
SWSAEKSIAQLNSNYGKKVLINFFVGTDDKNSMNHIIHIDQPRLGLPSRDYYECTGIYKE
ACTAYVDFMIAVAKLIRQEEGLPIDENQISVEMNKVMELEKEIANATTKSEDRNDPMLLY
NKMTLAQIQNNFSLEINGKPFSWSNFTNEIMSTVNINIPNEEDVVVYAPEYLIKLKPILT
KYFPRDFQNLFSWRFIMDLVSSLSRTYKDSRNAFRKALYGTTSESATWRRCANYVNGNME
NAVGRLYVEAAFAGESKHVVEDLIAQIREVFIQTLDDLTWMDAETKKKAEEKALAIKERI
GYPDDIVSNDNKLNNEYLELNYKEDEYFENIIQNLKFSQSKQLKKLREKVDKDEWITGAA
IVNAFYSSGRNQIVFPAGILQPPFFSAQQSNSLNYGGIGMVIGHEITHGFDDNGRNFNKD
GDLVDWWTQQSANNFKEQSQCMVYQYGNFSWDLAGGQHLNGINTLGENIADNGGIGQAYR
AYQNYVKKNGEEKLLPGIDLNHKQLFFLNFAQVWCGTYRPEYAVNSIKTDVHSPGNFRII
GSLQNSVEFSEAFQCPKNSYMNPEKKCRVW
|
|
|
|---|
| BDBM50050158 |
|---|
| n/a |
|---|
| Name | BDBM50050158 |
|---|
| Synonyms: | 1-[(S)-2-((R)-2-Mercapto-3-phenyl-propionylamino)-propionyl]-pyrrolidine-2-carboxylic acid | CHEMBL20010 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H22N2O4S |
|---|
| Mol. Mass. | 350.433 |
|---|
| SMILES | C[C@H](NC(=O)[C@H](S)Cc1ccccc1)C(=O)N1CCCC1C(O)=O |
|---|
| Structure | 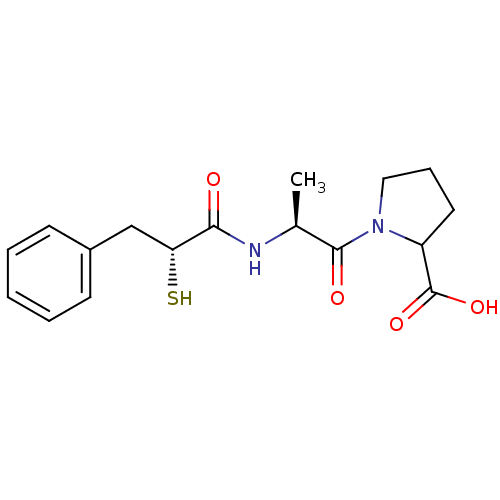
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Coric, P; Turcaud, S; Meudal, H; Roques, BP; Fournie-Zaluski, MC Optimal recognition of neutral endopeptidase and angiotensin-converting enzyme active sites by mercaptoacyldipeptides as a means to design potent dual inhibitors. J Med Chem39:1210-9 (1996) [PubMed] Article
Coric, P; Turcaud, S; Meudal, H; Roques, BP; Fournie-Zaluski, MC Optimal recognition of neutral endopeptidase and angiotensin-converting enzyme active sites by mercaptoacyldipeptides as a means to design potent dual inhibitors. J Med Chem39:1210-9 (1996) [PubMed] Article