

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Prothrombin | ||
| Ligand | BDBM50060709 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_208549 (CHEMBL814318) | ||
| Ki | 12.0±n/a nM | ||
| KON | 1800 M-1s-1 | ||
| Citation |  Tucker, TJ; Lumma, WC; Lewis, SD; Gardell, SJ; Lucas, BJ; Sisko, JT; Lynch, JJ; Lyle, EA; Baskin, EP; Woltmann, RF; Appleby, SD; Chen, IW; Dancheck, KB; Naylor-Olsen, AM; Krueger, JA; Cooper, CM; Vacca, JP Synthesis of a series of potent and orally bioavailable thrombin inhibitors that utilize 3,3-disubstituted propionic acid derivatives in the P3 position. J Med Chem40:3687-93 (1997) [PubMed] Article Tucker, TJ; Lumma, WC; Lewis, SD; Gardell, SJ; Lucas, BJ; Sisko, JT; Lynch, JJ; Lyle, EA; Baskin, EP; Woltmann, RF; Appleby, SD; Chen, IW; Dancheck, KB; Naylor-Olsen, AM; Krueger, JA; Cooper, CM; Vacca, JP Synthesis of a series of potent and orally bioavailable thrombin inhibitors that utilize 3,3-disubstituted propionic acid derivatives in the P3 position. J Med Chem40:3687-93 (1997) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Prothrombin | |||
| Name: | Prothrombin | ||
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain | ||
| Type: | Protein | ||
| Mol. Mass.: | 70029.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P00734 | ||
| Residue: | 622 | ||
| Sequence: |
| ||
| BDBM50060709 | |||
| n/a | |||
| Name | BDBM50060709 | ||
| Synonyms: | (S)-1-(3-Phenyl-3-pyridin-2-yl-propionyl)-pyrrolidine-2-carboxylic acid (4-amino-cyclohexylmethyl)-amide; TFA | CHEMBL332870 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H34N4O2 | ||
| Mol. Mass. | 434.5738 | ||
| SMILES | NC1CCC(CNC(=O)[C@@H]2CCCN2C(=O)CC(c2ccccc2)c2ccccn2)CC1 |wU:9.8,(6.95,-10.61,;5.99,-9.43,;5.61,-8.16,;6.25,-6.56,;4.9,-5.8,;3.53,-5.18,;3.37,-3.64,;1.96,-3.02,;.71,-3.93,;1.8,-1.5,;2.95,-.48,;2.31,.93,;.81,.77,;.49,-.74,;-.92,-1.34,;-1.08,-2.88,;-2.16,-.45,;-2.16,1.09,;-.85,1.85,;.49,1.09,;1.83,1.85,;1.83,3.39,;.49,4.16,;-.85,3.39,;-3.51,1.85,;-3.51,3.41,;-4.82,4.18,;-6.16,3.41,;-6.13,1.85,;-4.82,1.11,;5.32,-6.97,;4.68,-8.66,)| | ||
| Structure | 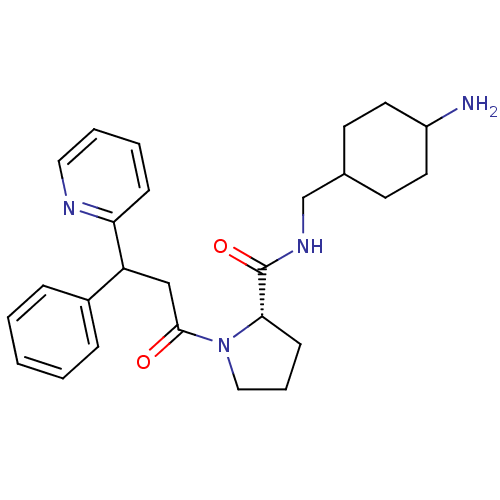
| ||