

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M3 | ||
| Ligand | BDBM50176731 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_327855 (CHEMBL863985) | ||
| Ki | 631±n/a nM | ||
| Citation |  Starck, JP; Talaga, P; Quéré, L; Collart, P; Christophe, B; Lo Brutto, P; Jadot, S; Chimmanamada, D; Zanda, M; Wagner, A; Mioskowski, C; Massingham, R; Guyaux, M Potent anti-muscarinic activity in a novel series of quinuclidine derivatives. Bioorg Med Chem Lett16:373-7 (2005) [PubMed] Article Starck, JP; Talaga, P; Quéré, L; Collart, P; Christophe, B; Lo Brutto, P; Jadot, S; Chimmanamada, D; Zanda, M; Wagner, A; Mioskowski, C; Massingham, R; Guyaux, M Potent anti-muscarinic activity in a novel series of quinuclidine derivatives. Bioorg Med Chem Lett16:373-7 (2005) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M3 | |||
| Name: | Muscarinic acetylcholine receptor M3 | ||
| Synonyms: | ACM3_HUMAN | CHRM3 | Cholinergic, muscarinic M3 | Muscarinic Receptors M3 | Muscarinic receptor M3 | RecName: Full=Muscarinic acetylcholine receptor M3 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 66151.03 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P20309 | ||
| Residue: | 590 | ||
| Sequence: |
| ||
| BDBM50176731 | |||
| n/a | |||
| Name | BDBM50176731 | ||
| Synonyms: | 2-benzyl-4-(3-methoxyquinuclidin-3-yl)-1-phenylbut-3-yn-2-ol | CHEMBL197973 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H29NO2 | ||
| Mol. Mass. | 375.5033 | ||
| SMILES | COC1(CN2CCC1CC2)C#CC(O)(Cc1ccccc1)Cc1ccccc1 |THB:1:2:5.6:9.8,(11.2,-14.77,;12.69,-15.16,;13.1,-16.65,;13.3,-18.03,;14.83,-17.37,;16.2,-18,;15.92,-16.61,;14.57,-16,;14.64,-14.37,;15.09,-15.48,;11.6,-17.05,;10.11,-17.44,;8.62,-17.84,;8.22,-16.35,;7.12,-18.24,;6.72,-19.72,;5.23,-20.11,;4.83,-21.6,;5.92,-22.69,;7.41,-22.29,;7.81,-20.8,;9.01,-19.33,;10.5,-19.73,;10.89,-21.22,;12.37,-21.62,;13.47,-20.54,;13.07,-19.04,;11.58,-18.65,)| | ||
| Structure | 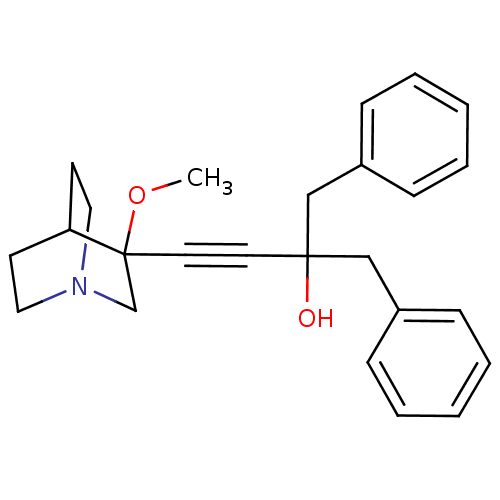
| ||