

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Beta-secretase 1 | ||
| Ligand | BDBM50372267 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_464832 (CHEMBL930550) | ||
| IC50 | 125±n/a nM | ||
| Citation |  Jennings, LD; Cole, DC; Stock, JR; Sukhdeo, MN; Ellingboe, JW; Cowling, R; Jin, G; Manas, ES; Fan, KY; Malamas, MS; Harrison, BL; Jacobsen, S; Chopra, R; Lohse, PA; Moore, WJ; O'Donnell, MM; Hu, Y; Robichaud, AJ; Turner, MJ; Wagner, E; Bard, J Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1' substrate binding pocket. Bioorg Med Chem Lett18:767-71 (2008) [PubMed] Article Jennings, LD; Cole, DC; Stock, JR; Sukhdeo, MN; Ellingboe, JW; Cowling, R; Jin, G; Manas, ES; Fan, KY; Malamas, MS; Harrison, BL; Jacobsen, S; Chopra, R; Lohse, PA; Moore, WJ; O'Donnell, MM; Hu, Y; Robichaud, AJ; Turner, MJ; Wagner, E; Bard, J Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1' substrate binding pocket. Bioorg Med Chem Lett18:767-71 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Beta-secretase 1 | |||
| Name: | Beta-secretase 1 | ||
| Synonyms: | ASP2 | Asp 2 | Aspartyl protease 2 | BACE | BACE1 | BACE1_HUMAN | Beta-secretase (BACE) | Beta-secretase 1 | Beta-secretase 1 (BACE 1) | Beta-secretase 1 (BACE-1) | Beta-secretase 1 (BACE1) | Beta-site APP cleaving enzyme 1 | Beta-site amyloid precursor protein cleaving enzyme 1 | KIAA1149 | Memapsin-2 | Membrane-associated aspartic protease 2 | beta-Secretase (BACE-1) | beta-Secretase (BACE1) | ||
| Type: | Protein | ||
| Mol. Mass.: | 55755.10 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P56817 | ||
| Residue: | 501 | ||
| Sequence: |
| ||
| BDBM50372267 | |||
| n/a | |||
| Name | BDBM50372267 | ||
| Synonyms: | CHEMBL428075 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H33ClN4O3 | ||
| Mol. Mass. | 509.04 | ||
| SMILES | CCCOc1ccc(cc1)-c1ccc(-c2ccccc2Cl)n1CC(=O)NC(=N)N[C@H]1CC[C@H](O)CC1 |wU:29.31,wD:32.35,(11.88,2.57,;11.9,1.03,;13.24,.28,;13.26,-1.26,;14.71,-1.78,;15.86,-.75,;17.32,-1.22,;17.63,-2.73,;16.5,-3.76,;15.04,-3.29,;19.1,-3.21,;19.58,-4.67,;21.12,-4.67,;21.59,-3.21,;23.06,-2.73,;23.37,-1.23,;24.83,-.75,;25.98,-1.78,;25.66,-3.29,;24.2,-3.76,;23.88,-5.27,;20.35,-2.3,;20.35,-.76,;21.68,.01,;23.01,-.76,;21.68,1.55,;23.01,2.32,;23.01,3.86,;24.35,1.55,;25.68,2.32,;25.67,3.85,;27.02,4.62,;28.35,3.85,;29.69,4.61,;28.34,2.3,;27.01,1.54,)| | ||
| Structure | 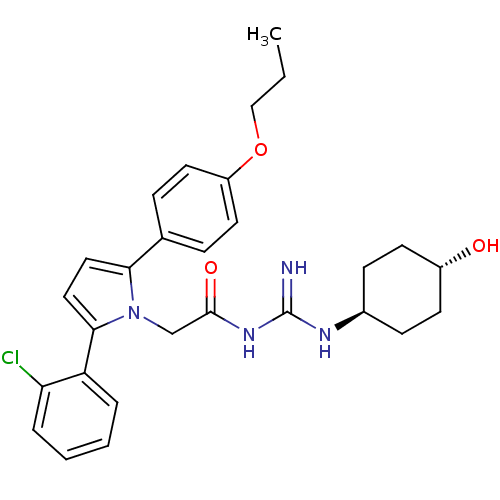
| ||