| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Muscarinic acetylcholine receptor M3 |
|---|
| Ligand | BDBM50212239 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_466555 (CHEMBL935647) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Jarvis, MF; Honore, P; Shieh, CC; Chapman, M; Joshi, S; Zhang, XF; Kort, M; Carroll, W; Marron, B; Atkinson, R; Thomas, J; Liu, D; Krambis, M; Liu, Y; McGaraughty, S; Chu, K; Roeloffs, R; Zhong, C; Mikusa, JP; Hernandez, G; Gauvin, D; Wade, C; Zhu, C; Pai, M; Scanio, M; Shi, L; Drizin, I; Gregg, R; Matulenko, M; Hakeem, A; Gross, M; Johnson, M; Marsh, K; Wagoner, PK; Sullivan, JP; Faltynek, CR; Krafte, DS A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci U S A104:8520-5 (2007) [PubMed] Article Jarvis, MF; Honore, P; Shieh, CC; Chapman, M; Joshi, S; Zhang, XF; Kort, M; Carroll, W; Marron, B; Atkinson, R; Thomas, J; Liu, D; Krambis, M; Liu, Y; McGaraughty, S; Chu, K; Roeloffs, R; Zhong, C; Mikusa, JP; Hernandez, G; Gauvin, D; Wade, C; Zhu, C; Pai, M; Scanio, M; Shi, L; Drizin, I; Gregg, R; Matulenko, M; Hakeem, A; Gross, M; Johnson, M; Marsh, K; Wagoner, PK; Sullivan, JP; Faltynek, CR; Krafte, DS A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci U S A104:8520-5 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Muscarinic acetylcholine receptor M3 |
|---|
| Name: | Muscarinic acetylcholine receptor M3 |
|---|
| Synonyms: | ACM3_HUMAN | CHRM3 | Cholinergic, muscarinic M3 | Muscarinic Receptors M3 | Muscarinic receptor M3 | RecName: Full=Muscarinic acetylcholine receptor M3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 66151.03 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P20309 |
|---|
| Residue: | 590 |
|---|
| Sequence: | MTLHNNSTTSPLFPNISSSWIHSPSDAGLPPGTVTHFGSYNVSRAAGNFSSPDGTTDDPL
GGHTVWQVVFIAFLTGILALVTIIGNILVIVSFKVNKQLKTVNNYFLLSLACADLIIGVI
SMNLFTTYIIMNRWALGNLACDLWLAIDYVASNASVMNLLVISFDRYFSITRPLTYRAKR
TTKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRTVPPGECFIQFLSEPTITFGTAIAAF
YMPVTIMTILYWRIYKETEKRTKELAGLQASGTEAETENFVHPTGSSRSCSSYELQQQSM
KRSNRRKYGRCHFWFTTKSWKPSSEQMDQDHSSSDSWNNNDAAASLENSASSDEEDIGSE
TRAIYSIVLKLPGHSTILNSTKLPSSDNLQVPEEELGMVDLERKADKLQAQKSVDDGGSF
PKSFSKLPIQLESAVDTAKTSDVNSSVGKSTATLPLSFKEATLAKRFALKTRSQITKRKR
MSLVKEKKAAQTLSAILLAFIITWTPYNIMVLVNTFCDSCIPKTFWNLGYWLCYINSTVN
PVCYALCNKTFRTTFKMLLLCQCDKKKRRKQQYQQRQSVIFHKRAPEQAL
|
|
|
|---|
| BDBM50212239 |
|---|
| n/a |
|---|
| Name | BDBM50212239 |
|---|
| Synonyms: | 5-(4-chlorophenyl)-N-(3,5-dimethoxyphenyl)furan-2-carboxamide | A-803467 | CHEMBL250699 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H16ClNO4 |
|---|
| Mol. Mass. | 357.788 |
|---|
| SMILES | COc1cc(NC(=O)c2ccc(o2)-c2ccc(Cl)cc2)cc(OC)c1 |
|---|
| Structure | 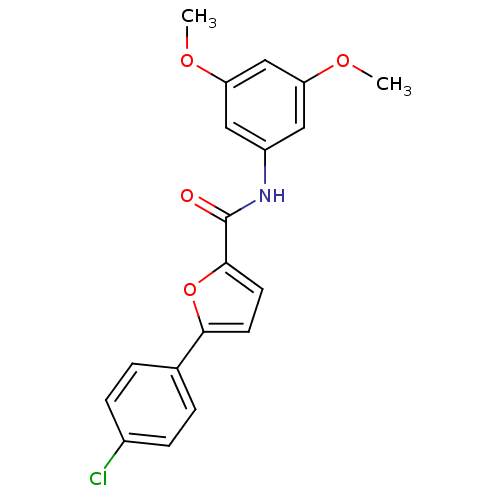
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jarvis, MF; Honore, P; Shieh, CC; Chapman, M; Joshi, S; Zhang, XF; Kort, M; Carroll, W; Marron, B; Atkinson, R; Thomas, J; Liu, D; Krambis, M; Liu, Y; McGaraughty, S; Chu, K; Roeloffs, R; Zhong, C; Mikusa, JP; Hernandez, G; Gauvin, D; Wade, C; Zhu, C; Pai, M; Scanio, M; Shi, L; Drizin, I; Gregg, R; Matulenko, M; Hakeem, A; Gross, M; Johnson, M; Marsh, K; Wagoner, PK; Sullivan, JP; Faltynek, CR; Krafte, DS A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci U S A104:8520-5 (2007) [PubMed] Article
Jarvis, MF; Honore, P; Shieh, CC; Chapman, M; Joshi, S; Zhang, XF; Kort, M; Carroll, W; Marron, B; Atkinson, R; Thomas, J; Liu, D; Krambis, M; Liu, Y; McGaraughty, S; Chu, K; Roeloffs, R; Zhong, C; Mikusa, JP; Hernandez, G; Gauvin, D; Wade, C; Zhu, C; Pai, M; Scanio, M; Shi, L; Drizin, I; Gregg, R; Matulenko, M; Hakeem, A; Gross, M; Johnson, M; Marsh, K; Wagoner, PK; Sullivan, JP; Faltynek, CR; Krafte, DS A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci U S A104:8520-5 (2007) [PubMed] Article