| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM50232053 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_468836 (CHEMBL947923) |
|---|
| IC50 | 11000±n/a nM |
|---|
| Citation |  Methot, JL; Chakravarty, PK; Chenard, M; Close, J; Cruz, JC; Dahlberg, WK; Fleming, J; Hamblett, CL; Hamill, JE; Harrington, P; Harsch, A; Heidebrecht, R; Hughes, B; Jung, J; Kenific, CM; Kral, AM; Meinke, PT; Middleton, RE; Ozerova, N; Sloman, DL; Stanton, MG; Szewczak, AA; Tyagarajan, S; Witter, DJ; Secrist, JP; Miller, TA Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg Med Chem Lett18:973-8 (2008) [PubMed] Article Methot, JL; Chakravarty, PK; Chenard, M; Close, J; Cruz, JC; Dahlberg, WK; Fleming, J; Hamblett, CL; Hamill, JE; Harrington, P; Harsch, A; Heidebrecht, R; Hughes, B; Jung, J; Kenific, CM; Kral, AM; Meinke, PT; Middleton, RE; Ozerova, N; Sloman, DL; Stanton, MG; Szewczak, AA; Tyagarajan, S; Witter, DJ; Secrist, JP; Miller, TA Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg Med Chem Lett18:973-8 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM50232053 |
|---|
| n/a |
|---|
| Name | BDBM50232053 |
|---|
| Synonyms: | CHEMBL271741 | N-(4-amino-biphenyl-3-yl)-benzamide | N-(4-aminobiphenyl-3-yl)benzamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H16N2O |
|---|
| Mol. Mass. | 288.3431 |
|---|
| SMILES | Nc1ccc(cc1NC(=O)c1ccccc1)-c1ccccc1 |
|---|
| Structure | 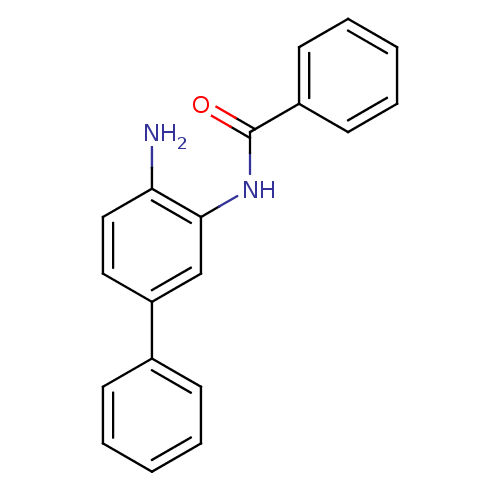
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Methot, JL; Chakravarty, PK; Chenard, M; Close, J; Cruz, JC; Dahlberg, WK; Fleming, J; Hamblett, CL; Hamill, JE; Harrington, P; Harsch, A; Heidebrecht, R; Hughes, B; Jung, J; Kenific, CM; Kral, AM; Meinke, PT; Middleton, RE; Ozerova, N; Sloman, DL; Stanton, MG; Szewczak, AA; Tyagarajan, S; Witter, DJ; Secrist, JP; Miller, TA Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg Med Chem Lett18:973-8 (2008) [PubMed] Article
Methot, JL; Chakravarty, PK; Chenard, M; Close, J; Cruz, JC; Dahlberg, WK; Fleming, J; Hamblett, CL; Hamill, JE; Harrington, P; Harsch, A; Heidebrecht, R; Hughes, B; Jung, J; Kenific, CM; Kral, AM; Meinke, PT; Middleton, RE; Ozerova, N; Sloman, DL; Stanton, MG; Szewczak, AA; Tyagarajan, S; Witter, DJ; Secrist, JP; Miller, TA Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg Med Chem Lett18:973-8 (2008) [PubMed] Article