| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor 3 |
|---|
| Ligand | BDBM50252922 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_539482 (CHEMBL1029767) |
|---|
| EC50 | 120000±n/a nM |
|---|
| Citation |  Butini, S; Pickering, DS; Morelli, E; Coccone, SS; Trotta, F; De Angelis, M; Guarino, E; Fiorini, I; Campiani, G; Novellino, E; Schousboe, A; Christensen, JK; Gemma, S 1H-cyclopentapyrimidine-2,4(1H,3H)-dione-related ionotropic glutamate receptors ligands. structure-activity relationships and identification of potent and Selective iGluR5 modulators. J Med Chem51:6614-8 (2008) [PubMed] Article Butini, S; Pickering, DS; Morelli, E; Coccone, SS; Trotta, F; De Angelis, M; Guarino, E; Fiorini, I; Campiani, G; Novellino, E; Schousboe, A; Christensen, JK; Gemma, S 1H-cyclopentapyrimidine-2,4(1H,3H)-dione-related ionotropic glutamate receptors ligands. structure-activity relationships and identification of potent and Selective iGluR5 modulators. J Med Chem51:6614-8 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor 3 |
|---|
| Name: | Glutamate receptor 3 |
|---|
| Synonyms: | GRIA3_RAT | Glur3 | Glutamate receptor 3 | Glutamate receptor ionotropic, AMPA 3 | Glutamate-AMPA | Gria3 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 100383.57 |
|---|
| Organism: | RAT |
|---|
| Description: | Glutamate-AMPA GRIA3 RAT::P19492 |
|---|
| Residue: | 888 |
|---|
| Sequence: | MGQSVLRAVFFLVLGLLGHSHGGFPNTISIGGLFMRNTVQEHSAFRFAVQLYNTNQNTTE
KPFHLNYHVDHLDSSNSFSVTNAFCSQFSRGVYAIFGFYDQMSMNTLTSFCGALHTSFVT
PSFPTDADVQFVIQMRPALKGAILSLLSYYKWEKFVYLYDTERGFSVLQAIMEAAVQNNW
QVTARSVGNIKDVQEFRRIIEEMDRRQEKRYLIDCEVERINTILEQVVILGKHSRGYHYM
LANLGFTDILLERVMHGGANITGFQIVNNENPMVQQFIQRWVRLDEREFPEAKNAPLKYT
SALTHDAILVIAEAFRYLRRQRVDVSRRGSAGDCLANPAVPWSQGIDIERALKMVQVQGM
TGNIQFDTYGRRTNYTIDVYEMKVSGSRKAGYWNEYERFVPFSDQQISNDSSSSENRTIV
VTTILESPYVMYKKNHEQLEGNERYEGYCVDLAYEIAKHVRIKYKLSIVGDGKYGARDPE
TKIWNGMVGELVYGRADIAVAPLTITLVREEVIDFSKPFMSLGISIMIKKPQKSKPGVFS
FLDPLAYEIWMCIVFAYIGVSVVLFLVSRFSPYEWHLEDNNEEPRDPQSPPDPPNEFGIF
NSLWFSLGAFMQQGCDISPRSLSGRIVGGVWWFFTLIIISSYTANLAAFLTVERMVSPIE
SAEDLAKQTEIAYGTLDSGSTKEFFRRSKIAVYEKMWSYMKSAEPSVFTKTTADGVARVR
KSKGKFAFLLESTMNEYIEQRKPCDTMKVGGNLDSKGYGVATPKGSALGNAVNLAVLKLN
EQGLLDKLKNKWWYDKGECGSGGGDSKDKTSALSLSNVAGVFYILVGGLGLAMMVALIEF
CYKSRAESKRMKLTKNTQNFKPAPATNTQNYATYREGYNVYGTESVKI
|
|
|
|---|
| BDBM50252922 |
|---|
| n/a |
|---|
| Name | BDBM50252922 |
|---|
| Synonyms: | (S)-1-(2'-Amino-2'-carboxyethyl)thieno[3,2-d]pyrimidin-2,4-dione | CHEMBL492630 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H9N3O4S |
|---|
| Mol. Mass. | 255.25 |
|---|
| SMILES | N[C@@H](Cn1c2ccsc2c(=O)[nH]c1=O)C(O)=O |r| |
|---|
| Structure | 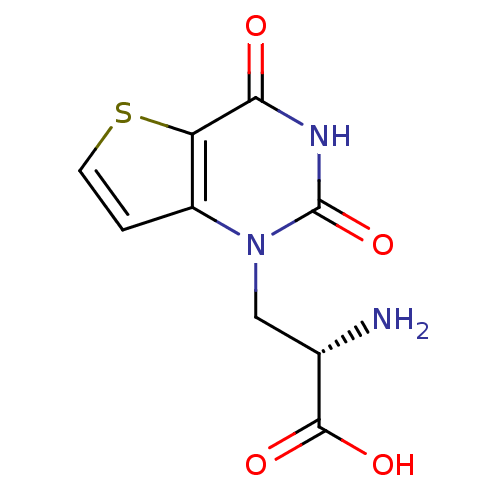
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Butini, S; Pickering, DS; Morelli, E; Coccone, SS; Trotta, F; De Angelis, M; Guarino, E; Fiorini, I; Campiani, G; Novellino, E; Schousboe, A; Christensen, JK; Gemma, S 1H-cyclopentapyrimidine-2,4(1H,3H)-dione-related ionotropic glutamate receptors ligands. structure-activity relationships and identification of potent and Selective iGluR5 modulators. J Med Chem51:6614-8 (2008) [PubMed] Article
Butini, S; Pickering, DS; Morelli, E; Coccone, SS; Trotta, F; De Angelis, M; Guarino, E; Fiorini, I; Campiani, G; Novellino, E; Schousboe, A; Christensen, JK; Gemma, S 1H-cyclopentapyrimidine-2,4(1H,3H)-dione-related ionotropic glutamate receptors ligands. structure-activity relationships and identification of potent and Selective iGluR5 modulators. J Med Chem51:6614-8 (2008) [PubMed] Article