

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Sodium/hydrogen exchanger 1 | ||
| Ligand | BDBM50297569 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_583577 (CHEMBL1059092) | ||
| IC50 | 12.2±n/a nM | ||
| Citation |  Zhang, R; Dong, J; Xu, YG; Hua, WY; Wen, N; You, QD Synthesis and bioactivity of substituted indan-1-ylideneaminoguanidine derivatives. Eur J Med Chem44:3771-6 (2009) [PubMed] Article Zhang, R; Dong, J; Xu, YG; Hua, WY; Wen, N; You, QD Synthesis and bioactivity of substituted indan-1-ylideneaminoguanidine derivatives. Eur J Med Chem44:3771-6 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Sodium/hydrogen exchanger 1 | |||
| Name: | Sodium/hydrogen exchanger 1 | ||
| Synonyms: | NHE-1 | Na(+)/H(+) exchanger 1 | Nhe1 | SL9A1_RAT | Slc9a1 | Solute carrier family 9 member 1 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 91655.00 | ||
| Organism: | Rattus norvegicus | ||
| Description: | ChEMBL_862370 | ||
| Residue: | 820 | ||
| Sequence: |
| ||
| BDBM50297569 | |||
| n/a | |||
| Name | BDBM50297569 | ||
| Synonyms: | CHEMBL563763 | N''-{5-chloro-2-[(5-nitro-1H-benzimidazol-2-yl)thio]-2,3-dihydro-1H-inden-1-ylidene}carbonohydrazonic diamide hydrochloride | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H14ClN7O2S | ||
| Mol. Mass. | 415.857 | ||
| SMILES | NC(N)=NN=C1C(Cc2cc(Cl)ccc12)Sc1nc2ccc(cc2[nH]1)[N+]([O-])=O |w:4.3,(6.38,-34.14,;4.87,-33.82,;4.39,-32.36,;3.84,-34.97,;2.33,-34.66,;1.3,-35.8,;-.23,-35.65,;-.86,-37.05,;.29,-38.08,;.29,-39.61,;1.62,-40.38,;1.62,-41.92,;2.95,-39.61,;2.95,-38.08,;1.63,-37.31,;-1,-34.32,;-2.54,-34.32,;-3.45,-35.58,;-4.93,-35.1,;-6.27,-35.87,;-7.6,-35.1,;-7.6,-33.55,;-6.28,-32.78,;-4.93,-33.54,;-3.46,-33.06,;-8.94,-32.78,;-8.95,-31.24,;-10.28,-33.55,)| | ||
| Structure | 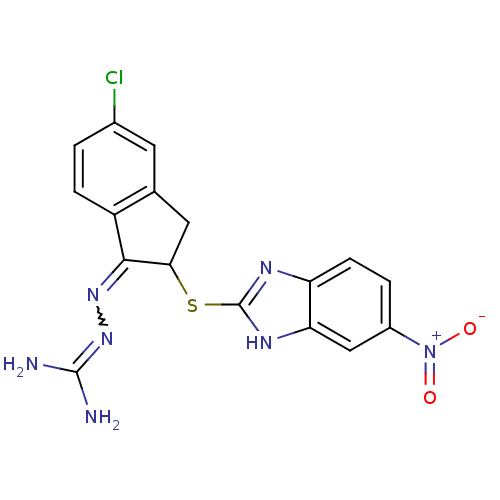
| ||