| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Orotidine 5'-phosphate decarboxylase |
|---|
| Ligand | BDBM50378784 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_639015 (CHEMBL1166153) |
|---|
| Temperature | 298.15±n/a K |
|---|
| Ki | 5±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Meza-Avina, ME; Wei, L; Liu, Y; Poduch, E; Bello, AM; Mishra, RK; Pai, EF; Kotra, LP Structural determinants for the inhibitory ligands of orotidine-5'-monophosphate decarboxylase. Bioorg Med Chem18:4032-41 (2010) [PubMed] Article Meza-Avina, ME; Wei, L; Liu, Y; Poduch, E; Bello, AM; Mishra, RK; Pai, EF; Kotra, LP Structural determinants for the inhibitory ligands of orotidine-5'-monophosphate decarboxylase. Bioorg Med Chem18:4032-41 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Orotidine 5'-phosphate decarboxylase |
|---|
| Name: | Orotidine 5'-phosphate decarboxylase |
|---|
| Synonyms: | Orotidine phosphate decarboxylase | PYRF_YEAST | URA3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 29241.18 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | ChEMBL_639015 |
|---|
| Residue: | 267 |
|---|
| Sequence: | MSKATYKERAATHPSPVAAKLFNIMHEKQTNLCASLDVRTTKELLELVEALGPKICLLKT
HVDILTDFSMEGTVKPLKALSAKYNFLLFEDRKFADIGNTVKLQYSAGVYRIAEWADITN
AHGVVGPGIVSGLKQAAEEVTKEPRGLLMLAELSCKGSLATGEYTKGTVDIAKSDKDFVI
GFIAQRDMGGRDEGYDWLIMTPGVGLDDKGDALGQQYRTVDDVVSTGSDIIIVGRGLFAK
GRDAKVEGERYRKAGWEAYLRRCGQQN
|
|
|
|---|
| BDBM50378784 |
|---|
| n/a |
|---|
| Name | BDBM50378784 |
|---|
| Synonyms: | CHEMBL1164953 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H14N3O9P |
|---|
| Mol. Mass. | 339.1959 |
|---|
| SMILES | NC(=O)c1[nH]nc([C@@H]2O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]2O)c1O |r| |
|---|
| Structure | 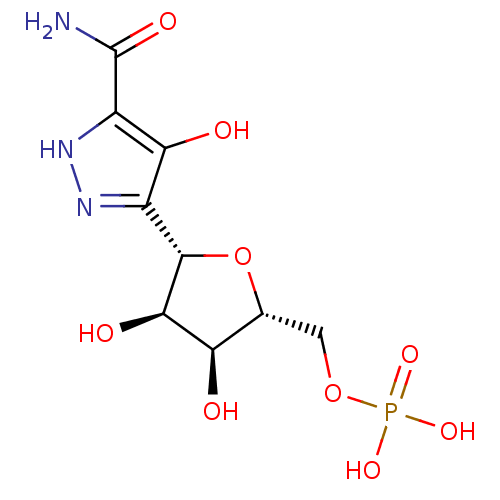
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Meza-Avina, ME; Wei, L; Liu, Y; Poduch, E; Bello, AM; Mishra, RK; Pai, EF; Kotra, LP Structural determinants for the inhibitory ligands of orotidine-5'-monophosphate decarboxylase. Bioorg Med Chem18:4032-41 (2010) [PubMed] Article
Meza-Avina, ME; Wei, L; Liu, Y; Poduch, E; Bello, AM; Mishra, RK; Pai, EF; Kotra, LP Structural determinants for the inhibitory ligands of orotidine-5'-monophosphate decarboxylase. Bioorg Med Chem18:4032-41 (2010) [PubMed] Article