

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Adenosine receptor A2a | ||
| Ligand | BDBM50393188 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_853052 (CHEMBL2154399) | ||
| IC50 | 263±n/a nM | ||
| Citation |  Drabczynska, A; Yuzlenko, O; Köse, M; Paskaleva, M; Schiedel, AC; Karolak-Wojciechowska, J; Handzlik, J; Karcz, T; Kuder, K; Müller, CE; Kiec-Kononowicz, K Synthesis and biological activity of tricyclic cycloalkylimidazo-, pyrimido- and diazepinopurinediones. Eur J Med Chem46:3590-607 (2011) [PubMed] Article Drabczynska, A; Yuzlenko, O; Köse, M; Paskaleva, M; Schiedel, AC; Karolak-Wojciechowska, J; Handzlik, J; Karcz, T; Kuder, K; Müller, CE; Kiec-Kononowicz, K Synthesis and biological activity of tricyclic cycloalkylimidazo-, pyrimido- and diazepinopurinediones. Eur J Med Chem46:3590-607 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Adenosine receptor A2a | |||
| Name: | Adenosine receptor A2a | ||
| Synonyms: | AA2AR_RAT | ADENOSINE A2a | Adenosine A2 receptor | Adenosine A2a receptor (A2a) | Adenosine Receptors A2a (A2a) | Adenosine receptor A2a and A3 | Adenosine receptors A2a | Adora2a | Rat striatal adenosine A2a receptor | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 45015.65 | ||
| Organism: | Rattus norvegicus (rat) | ||
| Description: | Rat A2A receptors expressed in CHO cells. | ||
| Residue: | 410 | ||
| Sequence: |
| ||
| BDBM50393188 | |||
| n/a | |||
| Name | BDBM50393188 | ||
| Synonyms: | CHEMBL2153620 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H25N5O2 | ||
| Mol. Mass. | 331.4127 | ||
| SMILES | CC1CCC(CC1)N1CCCn2c1nc1n(C)c(=O)n(C)c(=O)c21 |(38.63,-4.22,;37.75,-2.96,;36.21,-3.1,;35.33,-1.85,;35.97,-.45,;37.5,-.3,;38.39,-1.56,;35.09,.81,;35.73,2.22,;34.83,3.48,;33.29,3.33,;32.65,1.92,;33.55,.66,;32.65,-.58,;31.19,-.1,;29.86,-.86,;29.86,-2.4,;28.53,-.1,;27.2,-.87,;28.53,1.44,;27.2,2.2,;29.86,2.21,;29.86,3.75,;31.19,1.44,)| | ||
| Structure | 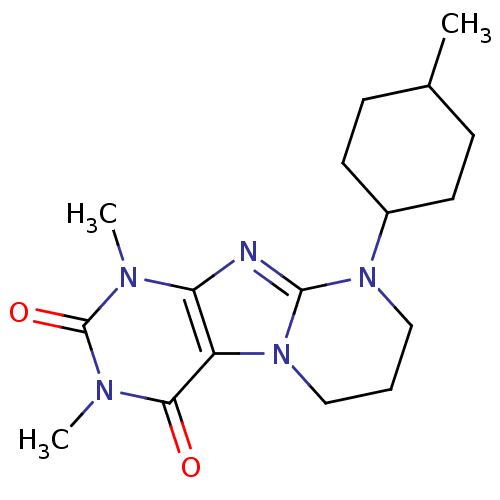
| ||