| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin E2 receptor EP1 subtype |
|---|
| Ligand | BDBM50422961 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_438068 (CHEMBL906324) |
|---|
| IC50 | 0.40±n/a nM |
|---|
| Citation |  Hall, A; Brown, SH; Chessell, IP; Chowdhury, A; Clayton, NM; Coleman, T; Giblin, GM; Hammond, B; Healy, MP; Johnson, MR; Metcalf, A; Michel, AD; Naylor, A; Novelli, R; Spalding, DJ; Sweeting, J; Winyard, L 1,5-Biaryl pyrrole derivatives as EP1 receptor antagonists. Structure-activity relationships of 6-substituted and 5,6-disubstituted benzoic acid derivatives. Bioorg Med Chem Lett17:916-20 (2007) [PubMed] Article Hall, A; Brown, SH; Chessell, IP; Chowdhury, A; Clayton, NM; Coleman, T; Giblin, GM; Hammond, B; Healy, MP; Johnson, MR; Metcalf, A; Michel, AD; Naylor, A; Novelli, R; Spalding, DJ; Sweeting, J; Winyard, L 1,5-Biaryl pyrrole derivatives as EP1 receptor antagonists. Structure-activity relationships of 6-substituted and 5,6-disubstituted benzoic acid derivatives. Bioorg Med Chem Lett17:916-20 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prostaglandin E2 receptor EP1 subtype |
|---|
| Name: | Prostaglandin E2 receptor EP1 subtype |
|---|
| Synonyms: | PE2R1_HUMAN | PGE receptor, EP1 subtype | PTGER1 | Prostaglandin E2 receptor | Prostaglandin E2 receptor EP1 subtype | Prostaglandin E2 receptor EP1 subtype (EP1) | Prostanoid EP1 receptor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 41834.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P34995 |
|---|
| Residue: | 402 |
|---|
| Sequence: | MSPCGPLNLSLAGEATTCAAPWVPNTSAVPPSGASPALPIFSMTLGAVSNLLALALLAQA
AGRLRRRRSAATFLLFVASLLATDLAGHVIPGALVLRLYTAGRAPAGGACHFLGGCMVFF
GLCPLLLGCGMAVERCVGVTRPLLHAARVSVARARLALAAVAAVALAVALLPLARVGRYE
LQYPGTWCFIGLGPPGGWRQALLAGLFASLGLVALLAALVCNTLSGLALLRARWRRRSRR
PPPASGPDSRRRWGAHGPRSASASSASSIASASTFFGGSRSSGSARRARAHDVEMVGQLV
GIMVVSCICWSPMLVLVALAVGGWSSTSLQRPLFLAVRLASWNQILDPWVYILLRQAVLR
QLLRLLPPRAGAKGGPAGLGLTPSAWEASSLRSSRHSGLSHF
|
|
|
|---|
| BDBM50422961 |
|---|
| n/a |
|---|
| Name | BDBM50422961 |
|---|
| Synonyms: | CHEMBL387969 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H21BrF2N2O4 |
|---|
| Mol. Mass. | 555.367 |
|---|
| SMILES | CC(=O)Nc1ccc(cc1C(O)=O)-n1c(C)ccc1-c1cc(Br)ccc1OCc1ccc(F)cc1F |
|---|
| Structure | 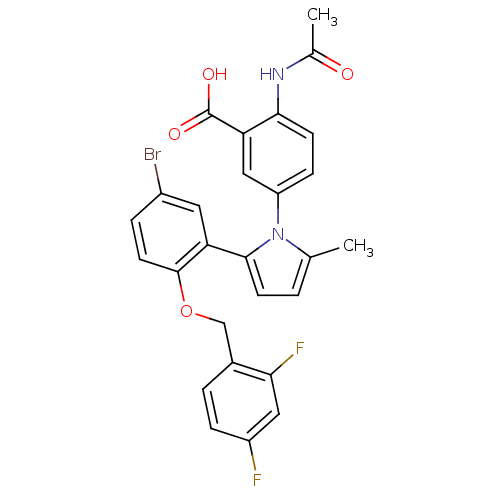
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hall, A; Brown, SH; Chessell, IP; Chowdhury, A; Clayton, NM; Coleman, T; Giblin, GM; Hammond, B; Healy, MP; Johnson, MR; Metcalf, A; Michel, AD; Naylor, A; Novelli, R; Spalding, DJ; Sweeting, J; Winyard, L 1,5-Biaryl pyrrole derivatives as EP1 receptor antagonists. Structure-activity relationships of 6-substituted and 5,6-disubstituted benzoic acid derivatives. Bioorg Med Chem Lett17:916-20 (2007) [PubMed] Article
Hall, A; Brown, SH; Chessell, IP; Chowdhury, A; Clayton, NM; Coleman, T; Giblin, GM; Hammond, B; Healy, MP; Johnson, MR; Metcalf, A; Michel, AD; Naylor, A; Novelli, R; Spalding, DJ; Sweeting, J; Winyard, L 1,5-Biaryl pyrrole derivatives as EP1 receptor antagonists. Structure-activity relationships of 6-substituted and 5,6-disubstituted benzoic acid derivatives. Bioorg Med Chem Lett17:916-20 (2007) [PubMed] Article