| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prostaglandin E synthase |
|---|
| Ligand | BDBM50426967 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_939016 (CHEMBL2328718) |
|---|
| IC50 | 3.0±n/a nM |
|---|
| Citation |  Walker, DP; Arhancet, GB; Lu, HF; Heasley, SE; Metz, S; Kablaoui, NM; Franco, FM; Hanau, CE; Scholten, JA; Springer, JR; Fobian, YM; Carter, JS; Xing, L; Yang, S; Shaffer, AF; Jerome, GM; Baratta, MT; Moore, WM; Vazquez, ML Synthesis and biological evaluation of substituted benzoxazoles as inhibitors of mPGES-1: use of a conformation-based hypothesis to facilitate compound design. Bioorg Med Chem Lett23:1120-6 (2013) [PubMed] Article Walker, DP; Arhancet, GB; Lu, HF; Heasley, SE; Metz, S; Kablaoui, NM; Franco, FM; Hanau, CE; Scholten, JA; Springer, JR; Fobian, YM; Carter, JS; Xing, L; Yang, S; Shaffer, AF; Jerome, GM; Baratta, MT; Moore, WM; Vazquez, ML Synthesis and biological evaluation of substituted benzoxazoles as inhibitors of mPGES-1: use of a conformation-based hypothesis to facilitate compound design. Bioorg Med Chem Lett23:1120-6 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prostaglandin E synthase |
|---|
| Name: | Prostaglandin E synthase |
|---|
| Synonyms: | MGST1L1 | MPGES1 | PGES | PIG12 | PTGES | PTGES_HUMAN | Prostaglandin E synthase (PGES-1) | Prostaglandin E synthase 1 (mPGES-1) | Prostaglandin E synthase-1 (PGES-1) | Prostaglandin E synthase/G/H synthase 2 | Prostaglandin E2 synthase-1 ( mPGES-1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 17112.22 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 152 |
|---|
| Sequence: | MPAHSLVMSSPALPAFLLCSTLLVIKMYVVAIITGQVRLRKKAFANPEDALRHGGPQYCR
SDPDVERCLRAHRNDMETIYPFLFLGFVYSFLGPNPFVAWMHFLVFLVGRVAHTVAYLGK
LRAPIRSVTYTLAQLPCASMALQILWEAARHL
|
|
|
|---|
| BDBM50426967 |
|---|
| n/a |
|---|
| Name | BDBM50426967 |
|---|
| Synonyms: | CHEMBL2325079 | PF-4693627 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H29Cl2N3O3 |
|---|
| Mol. Mass. | 502.433 |
|---|
| SMILES | OC[C@H]1CCC[C@@H](C1)NC(=O)C1CCN(CC1)c1nc2cc(Cl)c(cc2o1)-c1ccc(Cl)cc1 |r| |
|---|
| Structure | 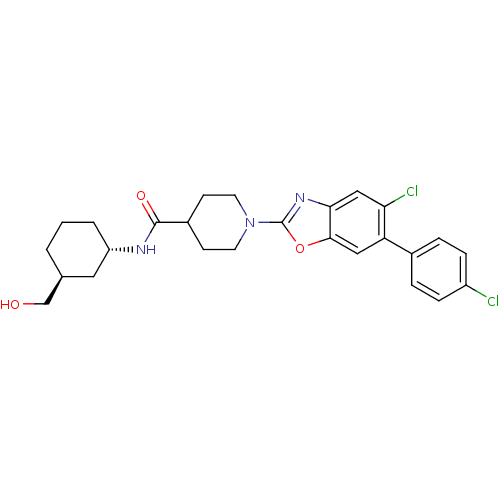
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Walker, DP; Arhancet, GB; Lu, HF; Heasley, SE; Metz, S; Kablaoui, NM; Franco, FM; Hanau, CE; Scholten, JA; Springer, JR; Fobian, YM; Carter, JS; Xing, L; Yang, S; Shaffer, AF; Jerome, GM; Baratta, MT; Moore, WM; Vazquez, ML Synthesis and biological evaluation of substituted benzoxazoles as inhibitors of mPGES-1: use of a conformation-based hypothesis to facilitate compound design. Bioorg Med Chem Lett23:1120-6 (2013) [PubMed] Article
Walker, DP; Arhancet, GB; Lu, HF; Heasley, SE; Metz, S; Kablaoui, NM; Franco, FM; Hanau, CE; Scholten, JA; Springer, JR; Fobian, YM; Carter, JS; Xing, L; Yang, S; Shaffer, AF; Jerome, GM; Baratta, MT; Moore, WM; Vazquez, ML Synthesis and biological evaluation of substituted benzoxazoles as inhibitors of mPGES-1: use of a conformation-based hypothesis to facilitate compound design. Bioorg Med Chem Lett23:1120-6 (2013) [PubMed] Article