

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Retinoic acid receptor RXR-alpha | ||
| Ligand | BDBM50429862 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_942867 (CHEMBL2342375) | ||
| EC50 | 367±n/a nM | ||
| Citation |  Ohsawa, F; Yamada, S; Yakushiji, N; Shinozaki, R; Nakayama, M; Kawata, K; Hagaya, M; Kobayashi, T; Kohara, K; Furusawa, Y; Fujiwara, C; Ohta, Y; Makishima, M; Naitou, H; Tai, A; Yoshikawa, Y; Yasui, H; Kakuta, H Mechanism of retinoid X receptor partial agonistic action of 1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)-1H-benzotriazole-5-carboxylic acid and structural development to increase potency. J Med Chem56:1865-77 (2013) [PubMed] Article Ohsawa, F; Yamada, S; Yakushiji, N; Shinozaki, R; Nakayama, M; Kawata, K; Hagaya, M; Kobayashi, T; Kohara, K; Furusawa, Y; Fujiwara, C; Ohta, Y; Makishima, M; Naitou, H; Tai, A; Yoshikawa, Y; Yasui, H; Kakuta, H Mechanism of retinoid X receptor partial agonistic action of 1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)-1H-benzotriazole-5-carboxylic acid and structural development to increase potency. J Med Chem56:1865-77 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Retinoic acid receptor RXR-alpha | |||
| Name: | Retinoic acid receptor RXR-alpha | ||
| Synonyms: | NR2B1 | Nuclear receptor subfamily 2 group B member 1 | Nuclear receptor subfamily 4 group A member 2 | Nuclear receptor subfamily 4 group A member 2/Retinoic acid receptor RXR-alpha | RXRA | RXRA_HUMAN | Retinoic acid receptor RXR-alpha/gamma | Retinoid X receptor alpha | Retinoid receptor | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 50820.38 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | ChEMBL_1456363 | ||
| Residue: | 462 | ||
| Sequence: |
| ||
| BDBM50429862 | |||
| n/a | |||
| Name | BDBM50429862 | ||
| Synonyms: | CHEMBL2146900 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C24H28N2O2 | ||
| Mol. Mass. | 376.4913 | ||
| SMILES | Cc1nc2cc(ccc2n1-c1cc2c(cc1C)C(C)(C)CCC2(C)C)C(O)=O |(13.72,1.99,;13.41,.48,;14.44,-.67,;13.67,-2.01,;14.15,-3.47,;13.12,-4.61,;11.61,-4.29,;11.13,-2.83,;12.16,-1.68,;12.01,-.16,;10.67,.61,;9.34,-.16,;8.01,.61,;8.01,2.15,;9.33,2.92,;10.66,2.16,;12,2.93,;6.67,2.93,;5.89,4.27,;7.44,4.27,;5.35,2.15,;5.35,.61,;6.67,-.16,;7.43,-1.49,;5.9,-1.48,;13.61,-6.08,;12.58,-7.23,;15.12,-6.4,)| | ||
| Structure | 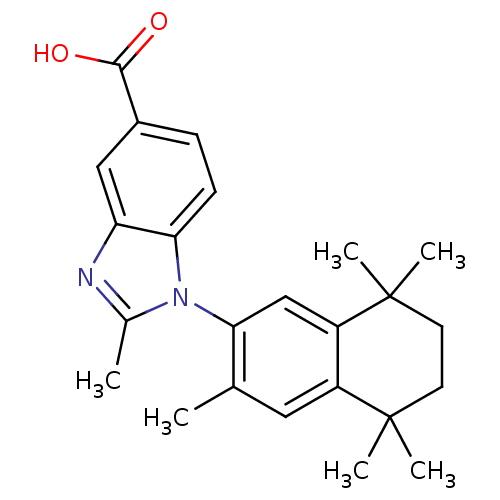
| ||