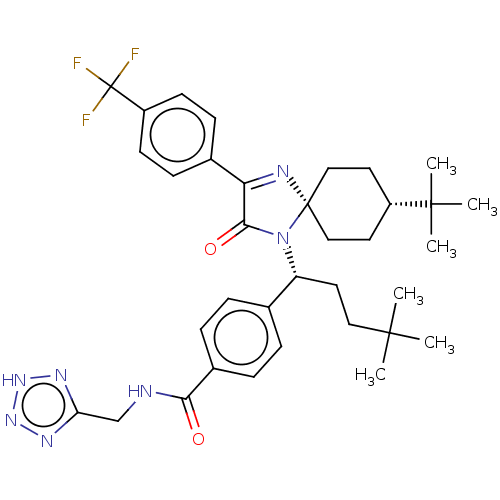
| Citation |  DeMong, D; Dai, X; Hwa, J; Miller, M; Lin, SI; Kang, L; Stamford, A; Greenlee, W; Yu, W; Wong, M; Lavey, B; Kozlowski, J; Zhou, G; Yang, DY; Patel, B; Soriano, A; Zhai, Y; Sondey, C; Zhang, H; Lachowicz, J; Grotz, D; Cox, K; Morrison, R; Andreani, T; Cao, Y; Liang, M; Meng, T; McNamara, P; Wong, J; Bradley, P; Feng, KI; Belani, J; Chen, P; Dai, P; Gauuan, J; Lin, P; Zhao, H The discovery of N-((2H-tetrazol-5-yl)methyl)-4-((R)-1-((5r,8R)-8-(tert-butyl)-3-(3,5-dichlorophenyl)-2-oxo-1,4-diazaspiro[4.5]dec-3-en-1-yl)-4,4-dimethylpentyl)benzamide (SCH 900822): a potent and selective glucagon receptor antagonist. J Med Chem57:2601-10 (2014) [PubMed] Article DeMong, D; Dai, X; Hwa, J; Miller, M; Lin, SI; Kang, L; Stamford, A; Greenlee, W; Yu, W; Wong, M; Lavey, B; Kozlowski, J; Zhou, G; Yang, DY; Patel, B; Soriano, A; Zhai, Y; Sondey, C; Zhang, H; Lachowicz, J; Grotz, D; Cox, K; Morrison, R; Andreani, T; Cao, Y; Liang, M; Meng, T; McNamara, P; Wong, J; Bradley, P; Feng, KI; Belani, J; Chen, P; Dai, P; Gauuan, J; Lin, P; Zhao, H The discovery of N-((2H-tetrazol-5-yl)methyl)-4-((R)-1-((5r,8R)-8-(tert-butyl)-3-(3,5-dichlorophenyl)-2-oxo-1,4-diazaspiro[4.5]dec-3-en-1-yl)-4,4-dimethylpentyl)benzamide (SCH 900822): a potent and selective glucagon receptor antagonist. J Med Chem57:2601-10 (2014) [PubMed] Article |
|---|
| SMILES | CC(C)(C)CC[C@@H](N1C(=O)C(=N[C@@]11CC[C@@H](CC1)C(C)(C)C)c1ccc(cc1)C(F)(F)F)c1ccc(cc1)C(=O)NCc1nn[nH]n1 |r,wU:15.19,6.5,wD:12.13,c:10,(32.97,-30.73,;32.95,-29.19,;31.61,-28.44,;31.61,-29.95,;34.28,-28.41,;34.26,-26.87,;35.58,-26.08,;35.53,-24.55,;36.74,-23.6,;38.22,-24.01,;36.21,-22.15,;34.66,-22.21,;34.25,-23.69,;33.06,-22.7,;31.61,-23.23,;31.35,-24.75,;32.54,-25.74,;33.99,-25.2,;29.91,-25.28,;28.73,-24.3,;28.46,-25.82,;29.65,-26.8,;36.98,-20.82,;38.53,-20.83,;39.3,-19.5,;38.54,-18.16,;37,-18.15,;36.22,-19.48,;39.32,-16.83,;40.86,-16.84,;38.56,-15.49,;40.08,-15.48,;36.91,-26.87,;36.88,-28.41,;38.21,-29.2,;39.56,-28.45,;39.58,-26.91,;38.25,-26.12,;40.88,-29.24,;40.86,-30.78,;42.22,-28.48,;43.54,-29.27,;44.89,-28.51,;46.28,-29.15,;47.33,-28.02,;46.57,-26.68,;45.06,-26.98,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 DeMong, D; Dai, X; Hwa, J; Miller, M; Lin, SI; Kang, L; Stamford, A; Greenlee, W; Yu, W; Wong, M; Lavey, B; Kozlowski, J; Zhou, G; Yang, DY; Patel, B; Soriano, A; Zhai, Y; Sondey, C; Zhang, H; Lachowicz, J; Grotz, D; Cox, K; Morrison, R; Andreani, T; Cao, Y; Liang, M; Meng, T; McNamara, P; Wong, J; Bradley, P; Feng, KI; Belani, J; Chen, P; Dai, P; Gauuan, J; Lin, P; Zhao, H The discovery of N-((2H-tetrazol-5-yl)methyl)-4-((R)-1-((5r,8R)-8-(tert-butyl)-3-(3,5-dichlorophenyl)-2-oxo-1,4-diazaspiro[4.5]dec-3-en-1-yl)-4,4-dimethylpentyl)benzamide (SCH 900822): a potent and selective glucagon receptor antagonist. J Med Chem57:2601-10 (2014) [PubMed] Article
DeMong, D; Dai, X; Hwa, J; Miller, M; Lin, SI; Kang, L; Stamford, A; Greenlee, W; Yu, W; Wong, M; Lavey, B; Kozlowski, J; Zhou, G; Yang, DY; Patel, B; Soriano, A; Zhai, Y; Sondey, C; Zhang, H; Lachowicz, J; Grotz, D; Cox, K; Morrison, R; Andreani, T; Cao, Y; Liang, M; Meng, T; McNamara, P; Wong, J; Bradley, P; Feng, KI; Belani, J; Chen, P; Dai, P; Gauuan, J; Lin, P; Zhao, H The discovery of N-((2H-tetrazol-5-yl)methyl)-4-((R)-1-((5r,8R)-8-(tert-butyl)-3-(3,5-dichlorophenyl)-2-oxo-1,4-diazaspiro[4.5]dec-3-en-1-yl)-4,4-dimethylpentyl)benzamide (SCH 900822): a potent and selective glucagon receptor antagonist. J Med Chem57:2601-10 (2014) [PubMed] Article