

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Voltage-dependent T-type calcium channel subunit alpha-1H | ||
| Ligand | BDBM50023589 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1454250 (CHEMBL3363800) | ||
| IC50 | >100000±n/a nM | ||
| Citation |  Reck, F; Ehmann, DE; Dougherty, TJ; Newman, JV; Hopkins, S; Stone, G; Agrawal, N; Ciaccio, P; McNulty, J; Barthlow, H; O'Donnell, J; Goteti, K; Breen, J; Comita-Prevoir, J; Cornebise, M; Cronin, M; Eyermann, CJ; Geng, B; Carr, GR; Pandarinathan, L; Tang, X; Cottone, A; Zhao, L; Bezdenejnih-Snyder, N Optimization of physicochemical properties and safety profile of novel bacterial topoisomerase type II inhibitors (NBTIs) with activity against Pseudomonas aeruginosa. Bioorg Med Chem22:5392-409 (2014) [PubMed] Article Reck, F; Ehmann, DE; Dougherty, TJ; Newman, JV; Hopkins, S; Stone, G; Agrawal, N; Ciaccio, P; McNulty, J; Barthlow, H; O'Donnell, J; Goteti, K; Breen, J; Comita-Prevoir, J; Cornebise, M; Cronin, M; Eyermann, CJ; Geng, B; Carr, GR; Pandarinathan, L; Tang, X; Cottone, A; Zhao, L; Bezdenejnih-Snyder, N Optimization of physicochemical properties and safety profile of novel bacterial topoisomerase type II inhibitors (NBTIs) with activity against Pseudomonas aeruginosa. Bioorg Med Chem22:5392-409 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Voltage-dependent T-type calcium channel subunit alpha-1H | |||
| Name: | Voltage-dependent T-type calcium channel subunit alpha-1H | ||
| Synonyms: | CAC1H_HUMAN | CACNA1H | Low-voltage-activated calcium channel alpha1 3.2 subunit | Voltage-gated T-type calcium channel | Voltage-gated calcium channel | Voltage-gated calcium channel subunit alpha Cav3.2 | voltage-dependent T-type calcium channel subunit alpha-1H isoform a | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 259187.68 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Calcium channel (Type T,L) 0 0::O95180 | ||
| Residue: | 2353 | ||
| Sequence: |
| ||
| BDBM50023589 | |||
| n/a | |||
| Name | BDBM50023589 | ||
| Synonyms: | CHEMBL3355573 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H26FN7O3 | ||
| Mol. Mass. | 467.496 | ||
| SMILES | [H][C@@]1(CC[C@@H](CC1)NCc1ncc2OCC(=O)Nc2n1)[C@@H](N)Cn1c2cc(F)ccc2ncc1=O |r,wU:20.23,wD:4.7,1.0,(25.26,-46.78,;23.93,-46.02,;22.6,-45.26,;22.59,-43.72,;23.93,-42.94,;25.26,-43.71,;25.27,-45.24,;23.92,-41.4,;25.26,-40.63,;26.59,-41.4,;26.58,-42.95,;27.92,-43.71,;29.25,-42.94,;30.58,-43.71,;31.92,-42.94,;31.92,-41.39,;33.25,-40.62,;30.58,-40.61,;29.24,-41.39,;27.91,-40.63,;23.94,-47.56,;25.28,-48.32,;22.61,-48.34,;22.62,-49.88,;21.29,-50.66,;19.95,-49.9,;18.62,-50.67,;17.29,-49.9,;18.62,-52.21,;19.96,-52.98,;21.29,-52.22,;22.63,-52.99,;23.98,-52.21,;23.97,-50.65,;25.3,-49.88,)| | ||
| Structure | 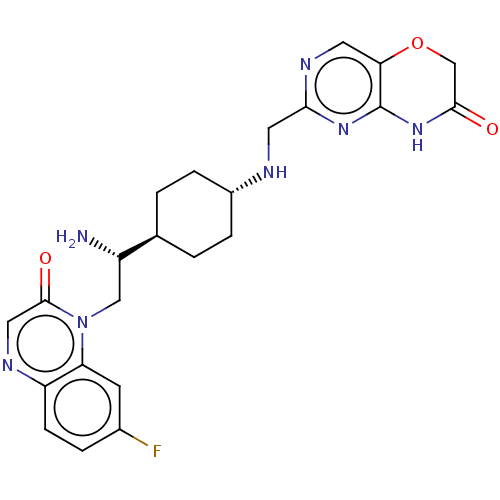
| ||