| Reaction Details |
|---|
 | Report a problem with these data |
| Target | N-acylethanolamine-hydrolyzing acid amidase |
|---|
| Ligand | BDBM50032457 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1440171 (CHEMBL3383170) |
|---|
| IC50 | 21±n/a nM |
|---|
| Citation |  Ponzano, S; Berteotti, A; Petracca, R; Vitale, R; Mengatto, L; Bandiera, T; Cavalli, A; Piomelli, D; Bertozzi, F; Bottegoni, G Synthesis, biological evaluation, and 3D QSAR study of 2-methyl-4-oxo-3-oxetanylcarbamic acid esters as N-acylethanolamine acid amidase (NAAA) inhibitors. J Med Chem57:10101-11 (2014) [PubMed] Article Ponzano, S; Berteotti, A; Petracca, R; Vitale, R; Mengatto, L; Bandiera, T; Cavalli, A; Piomelli, D; Bertozzi, F; Bottegoni, G Synthesis, biological evaluation, and 3D QSAR study of 2-methyl-4-oxo-3-oxetanylcarbamic acid esters as N-acylethanolamine acid amidase (NAAA) inhibitors. J Med Chem57:10101-11 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| N-acylethanolamine-hydrolyzing acid amidase |
|---|
| Name: | N-acylethanolamine-hydrolyzing acid amidase |
|---|
| Synonyms: | ASAH-like protein | Asahl | N-acylethanolamine acid amidase (NAAA) | N-acylethanolamine-hydrolyzing acid amidase | N-acylethanolamine-hydrolyzing acid amidase (NAAA) | N-acylethanolamine-hydrolyzing acid amidase subunit alpha | N-acylethanolamine-hydrolyzing acid amidase subunit beta | N-acylsphingosine amidohydrolase-like | NAAA_RAT | Naaa |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 40306.53 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | Q5KTC7 |
|---|
| Residue: | 362 |
|---|
| Sequence: | MGTPAIRAACHGAHLALALLLLLSLSDPWLWATAPGTPPLFNVSLDAAPELRWLPMLQHY
DPDFVRAAVAQVIGDRVPQWILEMIGEIVQKVESFLPQPFTSEIRGICDYLNLSLAEGVL
VNLAYEASAFCTSIVAQDSQGRIYHGRNLDYPFGNALRKLTADVQFVKNGQIVFTATTFV
GYVGLWTGQSPHKFTISGDERDKGWWWENMIAALSLGHSPISWLIRKTLTESEDFEAAVY
TLAKTPLIADVYYIVGGTSPQEGVVITRDRGGPADIWPLDPLNGAWFRVETNYDHWEPVP
KRDDRRTPAIKALNATGQAHLSLETLFQVLSVFPVYNNYTIYTTVMSAAEPDKYMTMIRN
PS
|
|
|
|---|
| BDBM50032457 |
|---|
| n/a |
|---|
| Name | BDBM50032457 |
|---|
| Synonyms: | CHEMBL3354143 | US9353075, 45 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H15NO4S |
|---|
| Mol. Mass. | 317.36 |
|---|
| SMILES | C[C@@H]1OC(=O)[C@@H]1NC(=O)OCc1ccc(cc1)-c1ccsc1 |r| |
|---|
| Structure | 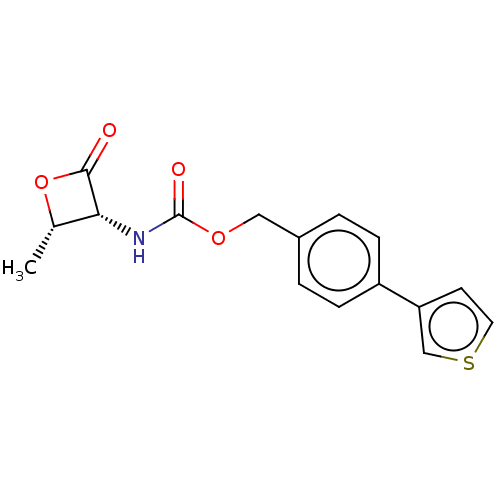
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ponzano, S; Berteotti, A; Petracca, R; Vitale, R; Mengatto, L; Bandiera, T; Cavalli, A; Piomelli, D; Bertozzi, F; Bottegoni, G Synthesis, biological evaluation, and 3D QSAR study of 2-methyl-4-oxo-3-oxetanylcarbamic acid esters as N-acylethanolamine acid amidase (NAAA) inhibitors. J Med Chem57:10101-11 (2014) [PubMed] Article
Ponzano, S; Berteotti, A; Petracca, R; Vitale, R; Mengatto, L; Bandiera, T; Cavalli, A; Piomelli, D; Bertozzi, F; Bottegoni, G Synthesis, biological evaluation, and 3D QSAR study of 2-methyl-4-oxo-3-oxetanylcarbamic acid esters as N-acylethanolamine acid amidase (NAAA) inhibitors. J Med Chem57:10101-11 (2014) [PubMed] Article